Structural basis for molecular recognition and presentation of histone H3 by WDR5
- PMID: 16946699
- PMCID: PMC1570438
- DOI: 10.1038/sj.emboj.7601316
Structural basis for molecular recognition and presentation of histone H3 by WDR5
Abstract
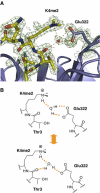
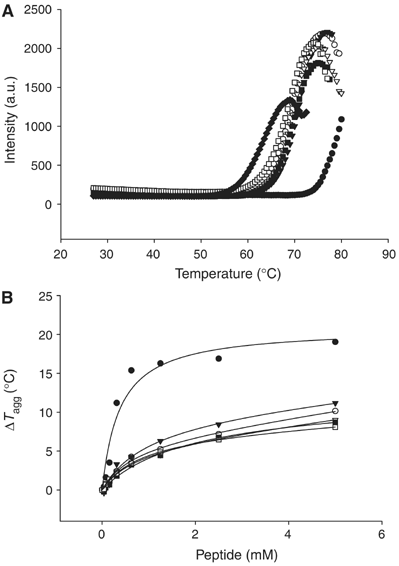
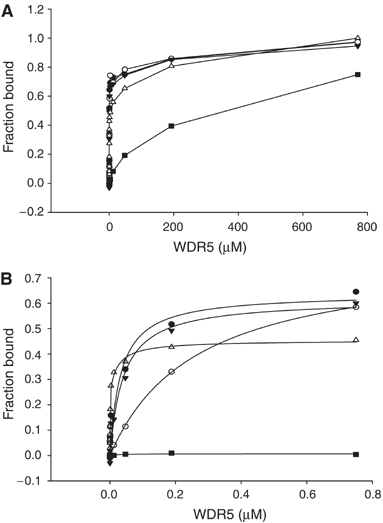
Histone methylation at specific lysine residues brings about various downstream events that are mediated by different effector proteins. The WD40 domain of WDR5 represents a new class of histone methyl-lysine recognition domains that is important for recruiting H3K4 methyltransferases to K4-dimethylated histone H3 tail as well as for global and gene-specific K4 trimethylation. Here we report the crystal structures of full-length WDR5, WDR5Delta23 and its complexes with unmodified, mono-, di- and trimethylated histone H3K4 peptides. The structures reveal that WDR5 is able to bind all of these histone H3 peptides, but only H3K4me2 peptide forms extra interactions with WDR5 by use of both water-mediated hydrogen bonding and the altered hydrophilicity of the modified lysine 4. We propose a mechanism for the involvement of WDR5 in binding and presenting histone H3K4 for further methylation as a component of MLL complexes.
Figures
Similar articles
-
Molecular recognition of histone H3 by the WD40 protein WDR5.Nat Struct Mol Biol. 2006 Aug;13(8):698-703. doi: 10.1038/nsmb1116. Epub 2006 Jul 9. Nat Struct Mol Biol. 2006. PMID: 16829960
-
Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex.Nat Struct Mol Biol. 2006 Aug;13(8):704-12. doi: 10.1038/nsmb1119. Epub 2006 Jul 9. Nat Struct Mol Biol. 2006. PMID: 16829959 Free PMC article.
-
Structural basis for the specific recognition of methylated histone H3 lysine 4 by the WD-40 protein WDR5.Mol Cell. 2006 Apr 7;22(1):137-44. doi: 10.1016/j.molcel.2006.03.018. Mol Cell. 2006. PMID: 16600877
-
On your histone mark, SET, methylate!Epigenetics. 2013 May;8(5):457-63. doi: 10.4161/epi.24451. Epub 2013 Apr 27. Epigenetics. 2013. PMID: 23625014 Free PMC article. Review.
-
On WD40 proteins: propelling our knowledge of transcriptional control?Epigenetics. 2012 Aug;7(8):815-22. doi: 10.4161/epi.21140. Epub 2012 Jul 19. Epigenetics. 2012. PMID: 22810296 Free PMC article. Review.
Cited by
-
Crystal Structure of the COMPASS H3K4 Methyltransferase Catalytic Module.Cell. 2018 Aug 23;174(5):1106-1116.e9. doi: 10.1016/j.cell.2018.06.038. Epub 2018 Aug 9. Cell. 2018. PMID: 30100181 Free PMC article.
-
Structural basis for the recognition of histone H4 by the histone-chaperone RbAp46.Structure. 2008 Jul;16(7):1077-85. doi: 10.1016/j.str.2008.05.006. Epub 2008 Jun 19. Structure. 2008. PMID: 18571423 Free PMC article.
-
The molecular biology of mixed lineage leukemia.Haematologica. 2009 Jul;94(7):984-93. doi: 10.3324/haematol.2008.002436. Epub 2009 Jun 16. Haematologica. 2009. PMID: 19535349 Free PMC article. Review.
-
Diverse functions of WD40 repeat proteins in histone recognition.Genes Dev. 2008 May 15;22(10):1265-8. doi: 10.1101/gad.1676208. Genes Dev. 2008. PMID: 18483215 Free PMC article.
-
CUL4A induces epithelial-mesenchymal transition and promotes cancer metastasis by regulating ZEB1 expression.Cancer Res. 2014 Jan 15;74(2):520-31. doi: 10.1158/0008-5472.CAN-13-2182. Epub 2013 Dec 4. Cancer Res. 2014. PMID: 24305877 Free PMC article.
References
-
- Chuikov S, Kurash JK, Wilson JR, Xiao B, Justin N, Ivanov GS, McKinney K, Tempst P, Prives C, Gamblin SJ, Barlev NA, Reinberg D (2004) Regulation of p53 activity through lysine methylation. Nature 432: 353–360 - PubMed
-
- Couture JF, Collazo E, Hauk G, Trievel RC (2006a) Structural basis for the methylation site specificity of SET7/9. Nat Struct Mol Biol 13: 140–146 - PubMed
-
- Couture JF, Collazo E, Hauk G, Trievel RC (2006b) Molecular recognition of histone H3 by the WD40 protein. Nat Struct Mol Biol 13: 698–703 - PubMed
-
- Dou Y, Milne TA, Tackett AJ, Smith ER, Fukuda A, Wysocka J, Allis CD, Chait BT, Hess JL, Roeder RG (2005) Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell 121: 873–885 - PubMed
-
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D 60: 2126–2132 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

