Specific recognition of ZNF217 and other zinc finger proteins at a surface groove of C-terminal binding proteins
- PMID: 16940172
- PMCID: PMC1636751
- DOI: 10.1128/MCB.00680-06
Specific recognition of ZNF217 and other zinc finger proteins at a surface groove of C-terminal binding proteins
Abstract
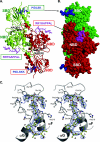
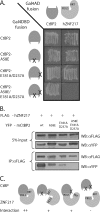
Numerous transcription factors recruit C-terminal binding protein (CtBP) corepressors. We show that the large zinc finger protein ZNF217 contacts CtBP. ZNF217 is encoded by an oncogene frequently amplified in tumors. ZNF217 contains a typical Pro-X-Asp-Leu-Ser (PXDLS) motif that binds in CtBP's PXDLS-binding cleft. However, ZNF217 also contains a second motif, Arg-Arg-Thr (RRT), that binds a separate surface on CtBP. The crystal structure of CtBP bound to an RRTGAPPAL peptide shows that it contacts a surface crevice distinct from the PXDLS binding cleft. Interestingly, both PXDLS and RRT motifs are also found in other zinc finger proteins, such as RIZ. Finally, we show that ZNF217 represses several promoters, including one from a known CtBP target gene, and mutations preventing ZNF217's contact with CtBP reduce repression. These results identify a new CtBP interaction motif and establish ZNF217 as a transcriptional repressor protein that functions, at least in part, by associating with CtBP.
Figures




Similar articles
-
Role of the C-terminal binding protein PXDLS motif binding cleft in protein interactions and transcriptional repression.Mol Cell Biol. 2006 Nov;26(21):8202-13. doi: 10.1128/MCB.00445-06. Epub 2006 Aug 28. Mol Cell Biol. 2006. PMID: 16940173 Free PMC article.
-
Human Krüppel-like factor 8: a CACCC-box binding protein that associates with CtBP and represses transcription.Nucleic Acids Res. 2000 May 1;28(9):1955-62. doi: 10.1093/nar/28.9.1955. Nucleic Acids Res. 2000. PMID: 10756197 Free PMC article.
-
The Ikaros family protein Eos associates with C-terminal-binding protein corepressors.Eur J Biochem. 2002 Dec;269(23):5885-92. doi: 10.1046/j.1432-1033.2002.03313.x. Eur J Biochem. 2002. PMID: 12444977
-
The CtBP family: enigmatic and enzymatic transcriptional co-repressors.Bioessays. 2001 Aug;23(8):683-90. doi: 10.1002/bies.1097. Bioessays. 2001. PMID: 11494316 Review.
-
Amplification of zinc finger gene 217 (ZNF217) and cancer: when good fingers go bad.Biochim Biophys Acta. 2007 Jun;1775(2):333-40. doi: 10.1016/j.bbcan.2007.05.001. Epub 2007 May 22. Biochim Biophys Acta. 2007. PMID: 17572303 Review.
Cited by
-
Interaction of CtBP with adenovirus E1A suppresses immortalization of primary epithelial cells and enhances virus replication during productive infection.Virology. 2013 Sep 1;443(2):313-20. doi: 10.1016/j.virol.2013.05.018. Epub 2013 Jun 5. Virology. 2013. PMID: 23747199 Free PMC article.
-
A role for Mediator complex subunit MED13L in Rb/E2F-induced growth arrest.Oncogene. 2012 Nov 1;31(44):4709-17. doi: 10.1038/onc.2011.622. Epub 2012 Jan 16. Oncogene. 2012. PMID: 22249253 Free PMC article.
-
Elevated expression of ZNF217 promotes prostate cancer growth by restraining ferroportin-conducted iron egress.Oncotarget. 2016 Dec 20;7(51):84893-84906. doi: 10.18632/oncotarget.12753. Oncotarget. 2016. PMID: 27768596 Free PMC article.
-
KDM1 class flavin-dependent protein lysine demethylases.Biopolymers. 2015 Jul;104(4):213-46. doi: 10.1002/bip.22643. Biopolymers. 2015. PMID: 25787087 Free PMC article. Review.
-
ZFP217 regulates adipogenesis by controlling mitotic clonal expansion in a METTL3-m6A dependent manner.RNA Biol. 2019 Dec;16(12):1785-1793. doi: 10.1080/15476286.2019.1658508. Epub 2019 Aug 27. RNA Biol. 2019. PMID: 31434544 Free PMC article.
References
-
- Chinnadurai, G. 2002. CtBP, an unconventional transcriptional corepressor in development and oncogenesis. Mol. Cell 9:213-224. - PubMed
-
- Collaborative Computational Project, no. 4. 1994. The CCP4 suite. Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760-763. - PubMed
-
- Collins, C., J. M. Rommens, D. Kowbel, T. Godfrey, M. Tanner, S. I. Hwang, D. Polikoff, G. Nonet, J. Cochran, K. Myambo, K. E. Jay, J. Froula, T. Cloutier, W. L. Kuo, P. Yaswen, S. Dairkee, J. Giovanola, G. B. Hutchinson, J. Isola, O. P. Kallioniemi, M. Palazzolo, C. Martin, C. Ericsson, D. Pinkel, D. Albertson, W. B. Li, and J. W. Gray. 1998. Positional cloning of ZNF217 and NABC1: genes amplified at 20q13.2 and overexpressed in breast carcinoma. Proc. Natl. Acad. Sci. USA 95:8703-8708. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
