Exopolysaccharide-associated protein sorting in environmental organisms: the PEP-CTERM/EpsH system. Application of a novel phylogenetic profiling heuristic
- PMID: 16930487
- PMCID: PMC1569441
- DOI: 10.1186/1741-7007-4-29
Exopolysaccharide-associated protein sorting in environmental organisms: the PEP-CTERM/EpsH system. Application of a novel phylogenetic profiling heuristic
Abstract
Background: Protein translocation to the proper cellular destination may be guided by various classes of sorting signals recognizable in the primary sequence. Detection in some genomes, but not others, may reveal sorting system components by comparison of the phylogenetic profile of the class of sorting signal to that of various protein families.
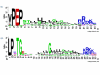
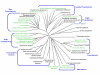
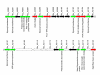
Results: We describe a short C-terminal homology domain, sporadically distributed in bacteria, with several key characteristics of protein sorting signals. The domain includes a near-invariant motif Pro-Glu-Pro (PEP). This possible recognition or processing site is followed by a predicted transmembrane helix and a cluster rich in basic amino acids. We designate this domain PEP-CTERM. It tends to occur multiple times in a genome if it occurs at all, with a median count of eight instances; Verrucomicrobium spinosum has sixty-five. PEP-CTERM-containing proteins generally contain an N-terminal signal peptide and exhibit high diversity and little homology to known proteins. All bacteria with PEP-CTERM have both an outer membrane and exopolysaccharide (EPS) production genes. By a simple heuristic for screening phylogenetic profiles in the absence of pre-formed protein families, we discovered that a homolog of the membrane protein EpsH (exopolysaccharide locus protein H) occurs in a species when PEP-CTERM domains are found. The EpsH family contains invariant residues consistent with a transpeptidase function. Most PEP-CTERM proteins are encoded by single-gene operons preceded by large intergenic regions. In the Proteobacteria, most of these upstream regions share a DNA sequence, a probable cis-regulatory site that contains a sigma-54 binding motif. The phylogenetic profile for this DNA sequence exactly matches that of three proteins: a sigma-54-interacting response regulator (PrsR), a transmembrane histidine kinase (PrsK), and a TPR protein (PrsT).
Conclusion: These findings are consistent with the hypothesis that PEP-CTERM and EpsH form a protein export sorting system, analogous to the LPXTG/sortase system of Gram-positive bacteria, and correlated to EPS expression. It occurs preferentially in bacteria from sediments, soils, and biofilms. The novel method that led to these findings, partial phylogenetic profiling, requires neither global sequence clustering nor arbitrary similarity cutoffs and appears to be a rapid, effective alternative to other profiling methods.
Figures

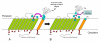

Similar articles
-
GlyGly-CTERM and rhombosortase: a C-terminal protein processing signal in a many-to-one pairing with a rhomboid family intramembrane serine protease.PLoS One. 2011;6(12):e28886. doi: 10.1371/journal.pone.0028886. Epub 2011 Dec 14. PLoS One. 2011. PMID: 22194940 Free PMC article.
-
Long-chain N-acyl amino acid synthases are linked to the putative PEP-CTERM/exosortase protein-sorting system in Gram-negative bacteria.J Bacteriol. 2011 Oct;193(20):5707-15. doi: 10.1128/JB.05426-11. Epub 2011 Aug 12. J Bacteriol. 2011. PMID: 21840974 Free PMC article.
-
Novel protein domains and motifs in the marine planctomycete Rhodopirellula baltica.FEMS Microbiol Lett. 2004 Jul 15;236(2):333-40. doi: 10.1016/j.femsle.2004.06.007. FEMS Microbiol Lett. 2004. PMID: 15251216
-
Peptide signal molecules and bacteriocins in Gram-negative bacteria: a genome-wide in silico screening for peptides containing a double-glycine leader sequence and their cognate transporters.Peptides. 2004 Sep;25(9):1425-40. doi: 10.1016/j.peptides.2003.10.028. Peptides. 2004. PMID: 15374646 Review.
-
STAND, a class of P-loop NTPases including animal and plant regulators of programmed cell death: multiple, complex domain architectures, unusual phyletic patterns, and evolution by horizontal gene transfer.J Mol Biol. 2004 Oct 8;343(1):1-28. doi: 10.1016/j.jmb.2004.08.023. J Mol Biol. 2004. PMID: 15381417 Review.
Cited by
-
Characterization of Outer Membrane Proteome of Akkermansia muciniphila Reveals Sets of Novel Proteins Exposed to the Human Intestine.Front Microbiol. 2016 Jul 26;7:1157. doi: 10.3389/fmicb.2016.01157. eCollection 2016. Front Microbiol. 2016. PMID: 27507967 Free PMC article.
-
Role of vimA in cell surface biogenesis in Porphyromonas gingivalis.Microbiology (Reading). 2010 Jul;156(Pt 7):2180-2193. doi: 10.1099/mic.0.038331-0. Epub 2010 Apr 8. Microbiology (Reading). 2010. PMID: 20378652 Free PMC article.
-
TIGRFAMs and Genome Properties: tools for the assignment of molecular function and biological process in prokaryotic genomes.Nucleic Acids Res. 2007 Jan;35(Database issue):D260-4. doi: 10.1093/nar/gkl1043. Epub 2006 Dec 6. Nucleic Acids Res. 2007. PMID: 17151080 Free PMC article.
-
Modular Lipoprotein Toxins Transferred by Outer Membrane Exchange Target Discrete Cell Entry Pathways.mBio. 2021 Oct 26;12(5):e0238821. doi: 10.1128/mBio.02388-21. Epub 2021 Sep 14. mBio. 2021. PMID: 34517761 Free PMC article.
-
Cooperative Interaction of Janthinobacterium sp. SLB01 and Flavobacterium sp. SLB02 in the Diseased Sponge Lubomirskia baicalensis.Int J Mol Sci. 2020 Oct 30;21(21):8128. doi: 10.3390/ijms21218128. Int J Mol Sci. 2020. PMID: 33143227 Free PMC article.
References
-
- Pugsley AP. Processing and methylation of PuIG, a pilin-like component of the general secretory pathway of Klebsiella oxytoca. Mol Microbiol. 1993;9:295–308. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

