The unique C termini of orthopoxvirus gamma interferon binding proteins are essential for ligand binding
- PMID: 16928759
- PMCID: PMC1641743
- DOI: 10.1128/JVI.01015-06
The unique C termini of orthopoxvirus gamma interferon binding proteins are essential for ligand binding
Abstract
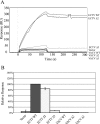
The orthopoxviruses ectromelia virus (ECTV) and vaccinia virus (VACV) express secreted gamma interferon binding proteins (IFN-gammaBPs) with homology to the ligand binding domains of the host's IFN-gamma receptor (IFN-gammaR1). Homology between these proteins is limited to the extracellular portions of the IFN-gammaR1 and the first approximately 200 amino acids of the IFN-gammaBPs. The remaining 60 amino acids at the C termini of the IFN-gammaBPs contain a single cysteine residue shown to be important in covalent dimerization of the secreted proteins. The function of the remaining C-terminal domain (CTD) has remained elusive, yet this region is conserved within all orthopoxvirus IFN-gammaBPs. Using a series of C-terminal deletion constructs, we have determined that the CTD is essential for IFN-gamma binding despite having no predicted homology to the IFN-gammaR1. Truncation of the ECTV IFN-gammaBP by more than two amino acid residues results in a complete loss of binding activity for both murine IFN-gamma and human IFN-gamma (hIFN-gamma), as measured by surface plasmon resonance (SPR) and bioassay. Equivalent truncation of the VACV IFN-gammaBP resulted in comparable loss of hIFN-gamma binding activity by SPR. Full-length IFN-gammaBPs were observed to form higher-ordered structures larger than the previously reported dimers. Mutants that were unable to bind IFN-gamma with high affinity in SPR experiments failed to assemble into these higher-ordered structures and migrated as dimers. We conclude that the unique CTD of orthopoxvirus IFN-gammaBPs is important for the assembly of covalent homodimers as well as the assembly of higher-ordered structures essential for IFN-gamma binding.
Figures



Similar articles
-
Identification of residues in the ectromelia virus gamma interferon-binding protein involved in expanded species specificity.J Gen Virol. 2007 Jan;88(Pt 1):51-60. doi: 10.1099/vir.0.82324-0. J Gen Virol. 2007. PMID: 17170436
-
Biosynthesis of the IFN-gamma binding protein of ectromelia virus, the causative agent of mousepox.Virology. 2005 Mar 30;334(1):41-50. doi: 10.1016/j.virol.2005.01.015. Virology. 2005. PMID: 15749121
-
Structure and mechanism of IFN-gamma antagonism by an orthopoxvirus IFN-gamma-binding protein.Proc Natl Acad Sci U S A. 2008 Feb 12;105(6):1861-6. doi: 10.1073/pnas.0705753105. Epub 2008 Feb 5. Proc Natl Acad Sci U S A. 2008. PMID: 18252829 Free PMC article.
-
Structural elements required for receptor recognition of human interferon-gamma.Pharmacol Ther. 1994 Oct;64(1):1-21. doi: 10.1016/0163-7258(94)90031-0. Pharmacol Ther. 1994. PMID: 7531344 Review.
-
The structure and function of interferon-gamma receptors.Int J Immunopharmacol. 1992 Apr;14(3):413-9. doi: 10.1016/0192-0561(92)90171-g. Int J Immunopharmacol. 1992. PMID: 1535615 Review.
Cited by
-
Orthopoxvirus Zoonoses-Do We Still Remember and Are Ready to Fight?Pathogens. 2023 Feb 21;12(3):363. doi: 10.3390/pathogens12030363. Pathogens. 2023. PMID: 36986285 Free PMC article. Review.
-
Poxvirus-encoded gamma interferon binding protein dampens the host immune response to infection.J Virol. 2007 Apr;81(7):3346-53. doi: 10.1128/JVI.01927-06. Epub 2007 Jan 17. J Virol. 2007. PMID: 17229697 Free PMC article.
-
Subversion of cytokine networks by virally encoded decoy receptors.Immunol Rev. 2012 Nov;250(1):199-215. doi: 10.1111/imr.12009. Immunol Rev. 2012. PMID: 23046131 Free PMC article. Review.
-
Poxvirus proteomics and virus-host protein interactions.Microbiol Mol Biol Rev. 2009 Dec;73(4):730-49. doi: 10.1128/MMBR.00026-09. Microbiol Mol Biol Rev. 2009. PMID: 19946139 Free PMC article. Review.
-
Mechanisms of immunomodulation by mammalian and viral decoy receptors: insights from structures.Nat Rev Immunol. 2017 Feb;17(2):112-129. doi: 10.1038/nri.2016.134. Epub 2016 Dec 28. Nat Rev Immunol. 2017. PMID: 28028310 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

