Methylation status of the Epstein-Barr virus (EBV) BamHI W latent cycle promoter and promoter activity: analysis with novel EBV-positive Burkitt and lymphoblastoid cell lines
- PMID: 16920819
- PMCID: PMC1641762
- DOI: 10.1128/JVI.01204-06
Methylation status of the Epstein-Barr virus (EBV) BamHI W latent cycle promoter and promoter activity: analysis with novel EBV-positive Burkitt and lymphoblastoid cell lines
Abstract
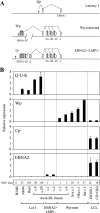
The Epstein-Barr virus (EBV) latent cycle promoter Wp, present in each tandemly arrayed copy of the BamHI W region in the EBV genome, drives expression of the EB viral nuclear antigens (EBNAs) at the initiation of virus-induced B-cell transformation. Thereafter, an alternative EBNA promoter, Cp, becomes dominant, Wp activity declines dramatically, and bisulfite sequencing of EBV-transformed lymphoblastoid cell lines (LCLs) shows extensive Wp methylation. Despite this, Wp is never completely silenced in LCLs. Here, using a combination of bisulfite sequencing and methylation-specific PCR, we show that in standard LCLs transformed with wild-type EBV isolates, some Wp copies always remain unmethylated, and in LCLs transformed with a recombinant EBV carrying just two BamHI W copies, Wp is completely unmethylated. Furthermore, we have analyzed rare LCLs, recently established using wild-type EBV isolates, and rare Burkitt lymphoma (BL) cell clones, recently established from tumors carrying EBNA2-deleted EBV genomes, which express EBNAs exclusively from Wp-initiated transcripts. Here, in sharp contrast to standard LCL and BL lines, all resident copies of Wp appear to be predominantly hypomethylated. Thus, studies of B cells with atypical patterns of Wp usage emphasize the strong correlation between the presence of unmethylated Wp sequences and promoter activity.
Figures





Similar articles
-
Variable methylation of the Epstein-Barr virus Wp EBNA gene promoter in B-lymphoblastoid cell lines.J Virol. 2004 Dec;78(24):14062-5. doi: 10.1128/JVI.78.24.14062-14065.2004. J Virol. 2004. PMID: 15564516 Free PMC article.
-
The Epstein-Barr virus (EBV) nuclear antigen 1 BamHI F promoter is activated on entry of EBV-transformed B cells into the lytic cycle.J Virol. 1992 Dec;66(12):7461-8. doi: 10.1128/JVI.66.12.7461-7468.1992. J Virol. 1992. PMID: 1331531 Free PMC article.
-
Methylation of transcription factor binding sites in the Epstein-Barr virus latent cycle promoter Wp coincides with promoter down-regulation during virus-induced B-cell transformation.J Virol. 2000 Nov;74(22):10468-79. doi: 10.1128/jvi.74.22.10468-10479.2000. J Virol. 2000. PMID: 11044091 Free PMC article.
-
Host cell-dependent expression of latent Epstein-Barr virus genomes: regulation by DNA methylation.Adv Cancer Res. 2003;89:133-56. doi: 10.1016/s0065-230x(03)01004-2. Adv Cancer Res. 2003. PMID: 14587872 Review.
-
Epigenetic regulation of latent Epstein-Barr virus promoters.Biochim Biophys Acta. 2010 Mar-Apr;1799(3-4):228-35. doi: 10.1016/j.bbagrm.2009.10.005. Epub 2009 Oct 22. Biochim Biophys Acta. 2010. PMID: 19853674 Review.
Cited by
-
Contributions of CTCF and DNA methyltransferases DNMT1 and DNMT3B to Epstein-Barr virus restricted latency.J Virol. 2012 Jan;86(2):1034-45. doi: 10.1128/JVI.05923-11. Epub 2011 Nov 9. J Virol. 2012. PMID: 22072770 Free PMC article.
-
The Role of Gammaherpesviruses in Cancer Pathogenesis.Pathogens. 2016 Feb 6;5(1):18. doi: 10.3390/pathogens5010018. Pathogens. 2016. PMID: 26861404 Free PMC article. Review.
-
trans-Repression of protein expression dependent on the Epstein-Barr virus promoter Wp during latency.J Virol. 2011 Nov;85(21):11435-47. doi: 10.1128/JVI.05158-11. Epub 2011 Aug 24. J Virol. 2011. PMID: 21865378 Free PMC article.
-
Epstein-Barr virus BamHI W repeat number limits EBNA2/EBNA-LP coexpression in newly infected B cells and the efficiency of B-cell transformation: a rationale for the multiple W repeats in wild-type virus strains.J Virol. 2011 Dec;85(23):12362-75. doi: 10.1128/JVI.06059-11. Epub 2011 Sep 28. J Virol. 2011. PMID: 21957300 Free PMC article.
-
Burkitt's lymphoma: the Rosetta Stone deciphering Epstein-Barr virus biology.Semin Cancer Biol. 2009 Dec;19(6):377-88. doi: 10.1016/j.semcancer.2009.07.004. Epub 2009 Jul 18. Semin Cancer Biol. 2009. PMID: 19619657 Free PMC article. Review.
References
-
- Alfieri, C., M. Birkenbach, and E. Kieff. 1991. Early events in Epstein-Barr virus infection of human B lymphocytes. Virology 181:595-608. - PubMed
-
- Allan, G. J., and D. T. Rowe. 1989. Size and stability of the Epstein-Barr virus major internal repeat (IR-1) in Burkitt's lymphoma and lymphoblastoid cell lines. Virology 173:489-498. - PubMed
-
- Allday, M. J., D. Kundu, S. Finerty, and B. E. Griffin. 1990. CpG methylation of viral DNA in EBV-associated tumours. Int. J. Cancer 45:1125-1130. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

