Highly conserved regions within the spike proteins of human coronaviruses 229E and NL63 determine recognition of their respective cellular receptors
- PMID: 16912312
- PMCID: PMC1563880
- DOI: 10.1128/JVI.00560-06
Highly conserved regions within the spike proteins of human coronaviruses 229E and NL63 determine recognition of their respective cellular receptors
Abstract
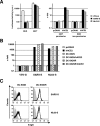
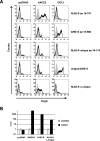
We have recently demonstrated that the severe acute respiratory syndrome coronavirus (SARS-CoV) receptor angiotensin converting enzyme 2 (ACE2) also mediates cellular entry of the newly discovered human coronavirus (hCoV) NL63. Here, we show that expression of DC-SIGN augments NL63 spike (S)-protein-driven infection of susceptible cells, while only expression of ACE2 but not DC-SIGN is sufficient for entry into nonpermissive cells, indicating that ACE2 fulfills the criteria of a bona fide hCoV-NL63 receptor. As for SARS-CoV, murine ACE2 is used less efficiently by NL63-S for entry than human ACE2. In contrast, several amino acid exchanges in human ACE2 which diminish SARS-S-driven entry do not interfere with NL63-S-mediated infection, suggesting that SARS-S and NL63-S might engage human ACE2 differentially. Moreover, we observed that NL63-S-driven entry was less dependent on a low-pH environment and activity of endosomal proteases compared to infection mediated by SARS-S, further suggesting differences in hCoV-NL63 and SARS-CoV cellular entry. NL63-S does not exhibit significant homology to SARS-S but is highly related to the S-protein of hCoV-229E, which enters target cells by engaging CD13. Employing mutagenic analyses, we found that the N-terminal unique domain in NL63-S, which is absent in 229E-S, does not confer binding to ACE2. In contrast, the highly homologous C-terminal parts of the NL63-S1 and 229E-S1 subunits in conjunction with distinct amino acids in the central regions of these proteins confer recognition of ACE2 and CD13, respectively. Therefore, despite the high homology of these sequences, they likely form sufficiently distinct surfaces, thus determining receptor specificity.
Figures

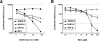



Similar articles
-
The S proteins of human coronavirus NL63 and severe acute respiratory syndrome coronavirus bind overlapping regions of ACE2.Virology. 2007 Oct 25;367(2):367-74. doi: 10.1016/j.virol.2007.04.035. Epub 2007 Jul 12. Virology. 2007. PMID: 17631932 Free PMC article.
-
Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry.Proc Natl Acad Sci U S A. 2005 May 31;102(22):7988-93. doi: 10.1073/pnas.0409465102. Epub 2005 May 16. Proc Natl Acad Sci U S A. 2005. PMID: 15897467 Free PMC article.
-
Differential downregulation of ACE2 by the spike proteins of severe acute respiratory syndrome coronavirus and human coronavirus NL63.J Virol. 2010 Jan;84(2):1198-205. doi: 10.1128/JVI.01248-09. Epub 2009 Oct 28. J Virol. 2010. PMID: 19864379 Free PMC article.
-
Properties of Coronavirus and SARS-CoV-2.Malays J Pathol. 2020 Apr;42(1):3-11. Malays J Pathol. 2020. PMID: 32342926 Review.
-
Severe acute respiratory syndrome coronavirus entry as a target of antiviral therapies.Antivir Ther. 2007;12(4 Pt B):639-50. Antivir Ther. 2007. PMID: 17944271 Review.
Cited by
-
Adaptive Evolution of the Spike Protein in Coronaviruses.Mol Biol Evol. 2023 Apr 4;40(4):msad089. doi: 10.1093/molbev/msad089. Mol Biol Evol. 2023. PMID: 37052956 Free PMC article.
-
Molecular mechanisms of human coronavirus NL63 infection and replication.Virus Res. 2023 Apr 2;327:199078. doi: 10.1016/j.virusres.2023.199078. Epub 2023 Feb 22. Virus Res. 2023. PMID: 36813239 Free PMC article. Review.
-
Design of immunogens for eliciting antibody responses that may protect against SARS-CoV-2 variants.PLoS Comput Biol. 2022 Sep 26;18(9):e1010563. doi: 10.1371/journal.pcbi.1010563. eCollection 2022 Sep. PLoS Comput Biol. 2022. PMID: 36156540 Free PMC article.
-
PEDV: Insights and Advances into Types, Function, Structure, and Receptor Recognition.Viruses. 2022 Aug 9;14(8):1744. doi: 10.3390/v14081744. Viruses. 2022. PMID: 36016366 Free PMC article. Review.
-
Multivalent ACE2 engineering-A promising pathway for advanced coronavirus nanomedicine development.Nano Today. 2022 Oct;46:101580. doi: 10.1016/j.nantod.2022.101580. Epub 2022 Aug 4. Nano Today. 2022. PMID: 35942040 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

