Mms2-Ubc13-dependent and -independent roles of Rad5 ubiquitin ligase in postreplication repair and translesion DNA synthesis in Saccharomyces cerevisiae
- PMID: 16908531
- PMCID: PMC1636848
- DOI: 10.1128/MCB.01260-06
Mms2-Ubc13-dependent and -independent roles of Rad5 ubiquitin ligase in postreplication repair and translesion DNA synthesis in Saccharomyces cerevisiae
Abstract
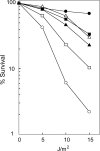
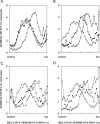
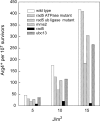
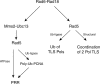
The Rad6-Rad18 ubiquitin-conjugating enzyme complex of Saccharomyces cerevisiae promotes replication through DNA lesions via three separate pathways that include translesion synthesis (TLS) by DNA polymerases eta and zeta and postreplicational repair (PRR) of discontinuities that form in the newly synthesized DNA opposite from DNA lesions, mediated by the Mms2-Ubc13 ubiquitin-conjugating enzyme and Rad5. Rad5 is an SWI/SNF family ATPase, and additionally, it functions as a ubiquitin ligase in the ubiquitin conjugation reaction. To decipher the roles of these Rad5 activities in lesion bypass, here we examine the effects of mutations in the Rad5 ATPase and ubiquitin ligase domains on the PRR of UV-damaged DNA and on UV-induced mutagenesis. Even though the ATPase-defective mutation confers only a modest degree of UV sensitivity whereas the ubiquitin ligase mutation causes a high degree of UV sensitivity, we find that both of these mutations produce the same high level of PRR defect as that conferred by the highly UV-sensitive rad5Delta mutation. From these studies, we infer a requirement of the Rad5 ATPase and ubiquitin ligase activities in PRR, and based upon the effects of different rad5 mutations on UV mutagenesis, we suggest a role for Rad5 in affecting the efficiency of lesion bypass by the TLS polymerases. In contrast to the role of Rad5 in PRR, however, where its function is coupled with that of Mms2-Ubc13, Rad5 function in TLS would be largely independent of this ubiquitin-conjugating enzyme complex.
Figures

Similar articles
-
Requirement of RAD5 and MMS2 for postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae.Mol Cell Biol. 2002 Apr;22(7):2419-26. doi: 10.1128/MCB.22.7.2419-2426.2002. Mol Cell Biol. 2002. PMID: 11884624 Free PMC article.
-
Opposing effects of ubiquitin conjugation and SUMO modification of PCNA on replicational bypass of DNA lesions in Saccharomyces cerevisiae.Mol Cell Biol. 2004 May;24(10):4267-74. doi: 10.1128/MCB.24.10.4267-4274.2004. Mol Cell Biol. 2004. PMID: 15121847 Free PMC article.
-
Requirement of Rad5 for DNA polymerase zeta-dependent translesion synthesis in Saccharomyces cerevisiae.Genetics. 2008 Sep;180(1):73-82. doi: 10.1534/genetics.108.091066. Epub 2008 Aug 30. Genetics. 2008. PMID: 18757916 Free PMC article.
-
Error-free DNA-damage tolerance in Saccharomyces cerevisiae.Mutat Res Rev Mutat Res. 2015 Apr-Jun;764:43-50. doi: 10.1016/j.mrrev.2015.02.001. Epub 2015 Feb 16. Mutat Res Rev Mutat Res. 2015. PMID: 26041265 Review.
-
Role of yeast Rad5 and its human orthologs, HLTF and SHPRH in DNA damage tolerance.DNA Repair (Amst). 2010 Mar 2;9(3):257-67. doi: 10.1016/j.dnarep.2009.12.013. Epub 2010 Jan 21. DNA Repair (Amst). 2010. PMID: 20096653 Review.
Cited by
-
HLTF's Ancient HIRAN Domain Binds 3' DNA Ends to Drive Replication Fork Reversal.Mol Cell. 2015 Jun 18;58(6):1090-100. doi: 10.1016/j.molcel.2015.05.013. Epub 2015 Jun 4. Mol Cell. 2015. PMID: 26051180 Free PMC article.
-
Concerted and differential actions of two enzymatic domains underlie Rad5 contributions to DNA damage tolerance.Nucleic Acids Res. 2015 Mar 11;43(5):2666-77. doi: 10.1093/nar/gkv004. Epub 2015 Feb 17. Nucleic Acids Res. 2015. PMID: 25690888 Free PMC article.
-
Control of DNA Damage Bypass by Ubiquitylation of PCNA.Genes (Basel). 2020 Jan 29;11(2):138. doi: 10.3390/genes11020138. Genes (Basel). 2020. PMID: 32013080 Free PMC article. Review.
-
Replicating damaged DNA in eukaryotes.Cold Spring Harb Perspect Biol. 2013 Dec 1;5(12):a019836. doi: 10.1101/cshperspect.a019836. Cold Spring Harb Perspect Biol. 2013. PMID: 24296172 Free PMC article. Review.
-
Involvement of budding yeast Rad5 in translesion DNA synthesis through physical interaction with Rev1.Nucleic Acids Res. 2016 Jun 20;44(11):5231-45. doi: 10.1093/nar/gkw183. Epub 2016 Mar 21. Nucleic Acids Res. 2016. PMID: 27001510 Free PMC article.
References
-
- Bailly, V., J. Lamb, P. Sung, S. Prakash, and L. Prakash. 1994. Specific complex formation between yeast RAD6 and RAD18 proteins: a potential mechanism for targeting RAD6 ubiquitin-conjugating activity to DNA damage sites. Genes Dev. 8:811-820. - PubMed
-
- Bailly, V., S. Lauder, S. Prakash, and L. Prakash. 1997. Yeast DNA repair proteins Rad6 and Rad18 form a heterodimer that has ubiquitin conjugating, DNA binding, and ATP hydrolytic activities. J. Biol. Chem. 272:23360-23365. - PubMed
-
- Bienko, M., C. M. Green, N. Crosetto, F. Rudolf, G. Zapart, B. Coull, P. Kannouche, G. Wider, M. Peter, A. R. Lehmann, K. Hofmann, and I. Dikic. 2005. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science 310:1821-1824. - PubMed
-
- Fang, S., J. P. Jensen, R. L. Ludwig, K. H. Vousden, and A. M. Weissman. 2000. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J. Biol. Chem. 275:8945-8951. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
