The cnidarian-bilaterian ancestor possessed at least 56 homeoboxes: evidence from the starlet sea anemone, Nematostella vectensis
- PMID: 16867185
- PMCID: PMC1779571
- DOI: 10.1186/gb-2006-7-7-R64
The cnidarian-bilaterian ancestor possessed at least 56 homeoboxes: evidence from the starlet sea anemone, Nematostella vectensis
Abstract
Background: Homeodomain transcription factors are key components in the developmental toolkits of animals. While this gene superclass predates the evolutionary split between animals, plants, and fungi, many homeobox genes appear unique to animals. The origin of particular homeobox genes may, therefore, be associated with the evolution of particular animal traits. Here we report the first near-complete set of homeodomains from a basal (diploblastic) animal.
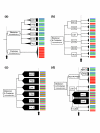
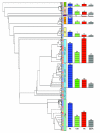
Results: Phylogenetic analyses were performed on 130 homeodomains from the sequenced genome of the sea anemone Nematostella vectensis along with 228 homeodomains from human and 97 homeodomains from Drosophila. The Nematostella homeodomains appear to be distributed among established homeodomain classes in the following fashion: 72 ANTP class; one HNF class; four LIM class; five POU class; 33 PRD class; five SINE class; and six TALE class. For four of the Nematostella homeodomains, there is disagreement between neighbor-joining and Bayesian trees regarding their class membership. A putative Nematostella CUT class gene is also identified.
Conclusion: The homeodomain superclass underwent extensive radiations prior to the evolutionary split between Cnidaria and Bilateria. Fifty-six homeodomain families found in human and/or fruit fly are also found in Nematostella, though seventeen families shared by human and fly appear absent in Nematostella. Homeodomain loss is also apparent in the bilaterian taxa: eight homeodomain families shared by Drosophila and Nematostella appear absent from human (CG13424, EMXLX, HOMEOBRAIN, MSXLX, NK7, REPO, ROUGH, and UNC4), and six homeodomain families shared by human and Nematostella appear absent from fruit fly (ALX, DMBX, DUX, HNF, POU1, and VAX).
Figures

Similar articles
-
Homeoboxes in sea anemones (Cnidaria:Anthozoa): a PCR-based survey of Nematostella vectensis and Metridium senile.Biol Bull. 1997 Aug;193(1):62-76. doi: 10.2307/1542736. Biol Bull. 1997. PMID: 9290214
-
Ancient complexity of the non-Hox ANTP gene complement in the anthozoan Nematostella vectensis: implications for the evolution of the ANTP superclass.J Exp Zool B Mol Dev Evol. 2006 Nov 15;306(6):589-96. doi: 10.1002/jez.b.21123. J Exp Zool B Mol Dev Evol. 2006. PMID: 16838293
-
A conserved cluster of three PRD-class homeobox genes (homeobrain, rx and orthopedia) in the Cnidaria and Protostomia.Evodevo. 2010 Jul 5;1(1):3. doi: 10.1186/2041-9139-1-3. Evodevo. 2010. PMID: 20849646 Free PMC article.
-
Environmental sensing and response genes in cnidaria: the chemical defensome in the sea anemone Nematostella vectensis.Cell Biol Toxicol. 2008 Dec;24(6):483-502. doi: 10.1007/s10565-008-9107-5. Epub 2008 Oct 28. Cell Biol Toxicol. 2008. PMID: 18956243 Free PMC article. Review.
-
Rising starlet: the starlet sea anemone, Nematostella vectensis.Bioessays. 2005 Feb;27(2):211-21. doi: 10.1002/bies.20181. Bioessays. 2005. PMID: 15666346 Review.
Cited by
-
Evolutionary origin and functional divergence of totipotent cell homeobox genes in eutherian mammals.BMC Biol. 2016 Jun 13;14:45. doi: 10.1186/s12915-016-0267-0. BMC Biol. 2016. PMID: 27296695 Free PMC article.
-
The homeodomain complement of the ctenophore Mnemiopsis leidyi suggests that Ctenophora and Porifera diverged prior to the ParaHoxozoa.Evodevo. 2010 Oct 4;1(1):9. doi: 10.1186/2041-9139-1-9. Evodevo. 2010. PMID: 20920347 Free PMC article.
-
Developmental expression of COE across the Metazoa supports a conserved role in neuronal cell-type specification and mesodermal development.Dev Genes Evol. 2010 Dec;220(7-8):221-34. doi: 10.1007/s00427-010-0343-3. Epub 2010 Nov 11. Dev Genes Evol. 2010. PMID: 21069538 Free PMC article.
-
The early ANTP gene repertoire: insights from the placozoan genome.PLoS One. 2008 Aug 21;3(8):e2457. doi: 10.1371/journal.pone.0002457. PLoS One. 2008. PMID: 18716659 Free PMC article.
-
Evolution of Prdm Genes in Animals: Insights from Comparative Genomics.Mol Biol Evol. 2016 Mar;33(3):679-96. doi: 10.1093/molbev/msv260. Epub 2015 Nov 11. Mol Biol Evol. 2016. PMID: 26560352 Free PMC article.
References
-
- DeRobertis EM. The homeobox in cell differentiation and evolution. In: Duboule D, editor. Guidebook to the Homeobox Genes. Oxford: Oxford University Press; 1994. pp. 13–23.
-
- Bürglin TR. A comprehensive classification of homeobox genes. In: Duboule D, editor. Guidebook to the Homeobox Genes. New York: Oxford University Press; 1994. pp. 25–72.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

