Complex interaction of Drosophila GRIP PDZ domains and Echinoid during muscle morphogenesis
- PMID: 16858411
- PMCID: PMC1538559
- DOI: 10.1038/sj.emboj.7601216
Complex interaction of Drosophila GRIP PDZ domains and Echinoid during muscle morphogenesis
Abstract
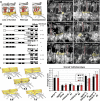
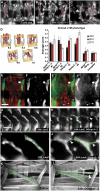
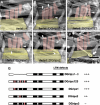
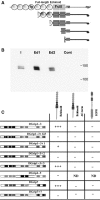
Glutamate receptor interacting protein (GRIP) homologues, initially characterized in synaptic glutamate receptor trafficking, consist of seven PDZ domains (PDZDs), whose conserved arrangement is of unknown significance. The Drosophila GRIP homologue (DGrip) is needed for proper guidance of embryonic somatic muscles towards epidermal attachment sites, with both excessive and reduced DGrip activity producing specific phenotypes in separate muscle groups. These phenotypes were utilized to analyze the molecular architecture underlying DGrip signaling function in vivo. Surprisingly, removing PDZDs 1-3 (DGripDelta1-3) or deleting ligand binding in PDZDs 1 or 2 convert DGrip to excessive in vivo activity mediated by ligand binding to PDZD 7. Yeast two-hybrid screening identifies the cell adhesion protein Echinoid's (Ed) type II PDZD-interaction motif as binding PDZDs 1, 2 and 7 of DGrip. ed loss-of-function alleles exhibit muscle defects, enhance defects caused by reduced DGrip activity and suppress the dominant DGripDelta1-3 effect during embryonic muscle formation. We propose that Ed and DGrip form a signaling complex, where competition between N-terminal and the C-terminal PDZDs of DGrip for Ed binding controls signaling function.
Figures



Similar articles
-
A glutamate receptor-interacting protein homolog organizes muscle guidance in Drosophila.Genes Dev. 2004 Jan 15;18(2):223-37. doi: 10.1101/gad.287604. Epub 2004 Jan 16. Genes Dev. 2004. PMID: 14729572 Free PMC article.
-
The transmembrane protein Perdido interacts with Grip and integrins to mediate myotube projection and attachment in the Drosophila embryo.Development. 2007 Dec;134(24):4469-78. doi: 10.1242/dev.014027. Development. 2007. PMID: 18039972
-
Nuclear trafficking of Drosophila Frizzled-2 during synapse development requires the PDZ protein dGRIP.Proc Natl Acad Sci U S A. 2006 May 16;103(20):7841-6. doi: 10.1073/pnas.0600387103. Epub 2006 May 8. Proc Natl Acad Sci U S A. 2006. PMID: 16682643 Free PMC article.
-
The many faces of cell adhesion during Drosophila muscle development.Dev Biol. 2015 May 1;401(1):62-74. doi: 10.1016/j.ydbio.2014.12.038. Epub 2015 Jan 14. Dev Biol. 2015. PMID: 25596335 Review.
-
PDZ proteins and polarity: functions from the fly.Trends Genet. 2001 Sep;17(9):511-9. doi: 10.1016/s0168-9525(01)02407-6. Trends Genet. 2001. PMID: 11525834 Review.
Cited by
-
Genetic Control of Muscle Diversification and Homeostasis: Insights from Drosophila.Cells. 2020 Jun 25;9(6):1543. doi: 10.3390/cells9061543. Cells. 2020. PMID: 32630420 Free PMC article. Review.
-
Hecate/Grip2a acts to reorganize the cytoskeleton in the symmetry-breaking event of embryonic axis induction.PLoS Genet. 2014 Jun 26;10(6):e1004422. doi: 10.1371/journal.pgen.1004422. eCollection 2014 Jun. PLoS Genet. 2014. PMID: 24967891 Free PMC article.
-
Drosophila importin-7 functions upstream of the Elmo signaling module to mediate the formation and stability of muscle attachments.J Cell Sci. 2013 Nov 15;126(Pt 22):5210-23. doi: 10.1242/jcs.132241. Epub 2013 Sep 17. J Cell Sci. 2013. PMID: 24046451 Free PMC article.
-
RacGAP50C directs perinuclear gamma-tubulin localization to organize the uniform microtubule array required for Drosophila myotube extension.Development. 2009 May;136(9):1411-21. doi: 10.1242/dev.031823. Epub 2009 Mar 18. Development. 2009. PMID: 19297411 Free PMC article.
-
Cooperation of Syd-1 with Neurexin synchronizes pre- with postsynaptic assembly.Nat Neurosci. 2012 Sep;15(9):1219-26. doi: 10.1038/nn.3183. Epub 2012 Aug 5. Nat Neurosci. 2012. PMID: 22864612
References
-
- Ahmed A, Chandra S, Magarinos M, Vaessin H (2003) Echinoid mutants exhibit neurogenic phenotypes and show synergistic interactions with the Notch signaling pathway. Development 130: 6295–6304 - PubMed
-
- Artero R, Furlong EE, Beckett K, Scott MP, Baylies M (2003) Notch and Ras signaling pathway effector genes expressed in fusion competent and founder cells during Drosophila myogenesis. Development 130: 6257–6272 - PubMed
-
- Bai J, Chiu W, Wang J, Tzeng T, Perrimon N, Hsu J (2001) The cell adhesion molecule Echinoid defines a new pathway that antagonizes the Drosophila EGF receptor signaling pathway. Development 128: 591–601 - PubMed
-
- Baran R, Jin Y (2002) Getting a GRIP on liprins. Neuron 34: 1–2 - PubMed
-
- Brand AH, Perrimon N (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

