A distant upstream locus control region is critical for expression of the Kit receptor gene in mast cells
- PMID: 16847336
- PMCID: PMC1592758
- DOI: 10.1128/MCB.01854-05
A distant upstream locus control region is critical for expression of the Kit receptor gene in mast cells
Abstract
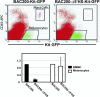
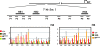
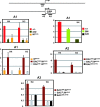
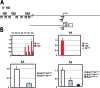
The Kit receptor tyrosine kinase functions in hematopoiesis, melanogenesis, and gametogenesis and in interstitial cells of Cajal. We previously identified two upstream hypersensitive site (HS) clusters in mast cells and melanocytes. Here we investigated the roles of these 5' HS sequences in Kit expression using transgenic mice carrying Kit-GFP reporter constructs. In these mice there is close correspondence between Kit-GFP reporter and endogenous Kit gene expression in most tissues analyzed. Deletion analysis defined the 5' upstream HS cluster region as critical for Kit expression in mast cells. Furthermore, chromatin immunoprecipitation analysis in mast cells showed that H3 and H4 histone hyperacetylation and RNA polymerase II recruitment within the Kit promoter and in the 5' HS region were associated with Kit expression. Therefore, the 5' upstream hypersensitivity sites appear to be critical components of locus control region-mediated Kit gene activation in mast cells.
Figures



Similar articles
-
Mast cell-deficient W-sash c-kit mutant Kit W-sh/W-sh mice as a model for investigating mast cell biology in vivo.Am J Pathol. 2005 Sep;167(3):835-48. doi: 10.1016/S0002-9440(10)62055-X. Am J Pathol. 2005. PMID: 16127161 Free PMC article.
-
Fyn kinase acts upstream of Shp2 and p38 mitogen-activated protein kinase to promote chemotaxis of mast cells towards stem cell factor.Cell Signal. 2006 Sep;18(9):1447-54. doi: 10.1016/j.cellsig.2005.11.005. Epub 2006 Jan 25. Cell Signal. 2006. PMID: 16442778
-
The W(sh), W(57), and Ph Kit expression mutations define tissue-specific control elements located between -23 and -154 kb upstream of Kit.Blood. 1999 Oct 15;94(8):2658-66. Blood. 1999. PMID: 10515869
-
The kit-ligand (steel factor) and its receptor c-kit/W: pleiotropic roles in gametogenesis and melanogenesis.Dev Suppl. 1993:125-37. Dev Suppl. 1993. PMID: 7519481 Review.
-
Regulation of gene expression in mast cells: micro-rNA expression and chromatin structural analysis of cytokine genes.Novartis Found Symp. 2005;271:179-87; discussion 187-90, 198-9. Novartis Found Symp. 2005. PMID: 16605135 Review.
Cited by
-
OCT4 and SOX2 Work as Transcriptional Activators in Reprogramming Human Fibroblasts.Cell Rep. 2017 Aug 15;20(7):1585-1596. doi: 10.1016/j.celrep.2017.07.071. Cell Rep. 2017. PMID: 28813671 Free PMC article.
-
Using c-kit to genetically target cerebellar molecular layer interneurons in adult mice.PLoS One. 2017 Jun 28;12(6):e0179347. doi: 10.1371/journal.pone.0179347. eCollection 2017. PLoS One. 2017. PMID: 28658323 Free PMC article.
-
Green fluorescent protein transgene driven by Kit regulatory sequences is expressed in hematopoietic stem cells.Haematologica. 2009 Mar;94(3):318-25. doi: 10.3324/haematol.13689. Epub 2009 Jan 30. Haematologica. 2009. PMID: 19181779 Free PMC article.
-
Polycomb Responds to Low Levels of Transcription.Cell Rep. 2017 Jul 25;20(4):785-793. doi: 10.1016/j.celrep.2017.06.076. Cell Rep. 2017. PMID: 28746865 Free PMC article.
-
Dendritic cell immunoreceptor drives atopic dermatitis by modulating oxidized CaMKII-involved mast cell activation.JCI Insight. 2022 Mar 8;7(5):e152559. doi: 10.1172/jci.insight.152559. JCI Insight. 2022. PMID: 35113811 Free PMC article.
References
-
- Agosti, V., S. Corbacioglu, I. Ehlers, C. Waskow, G. Sommer, G. Berrozpe, H. Kissel, C. M. Tucker, K. Manova, M. A. Moore, H. R. Rodewald, and P. Besmer. 2004. Critical role for Kit-mediated Src kinase but not PI 3-kinase signaling in pro T and pro B cell development. J. Exp. Med. 199:867-878. - PMC - PubMed
-
- Bachvarova, R. F., K. Manova, and P. Besmer. 1993. Role in gametogenesis of c-kit encoded at the W locus of mice. Wiley-Liss, New York, N.Y.
-
- Bedell, M. A., N. A. Jenkins, and N. G. Copeland. 1996. Good genes in bad neighbourhoods. Nat. Genet. 12:229-232. - PubMed
-
- Berrozpe, G., I. Timokhina, S. Yukl, Y. Tajima, M. Ono, A. D. Zelenetz, and P. Besmer. 1999. The W(sh), W(57), and Ph Kit expression mutations define tissue-specific control elements located between −23 and −154 kb upstream of Kit. Blood 94:2658-2666. - PubMed
-
- Besmer, P. 1997. Kit-ligand-stem cell factor. Marcel Dekker, New York, N.Y.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
