TFIIA plays a role in the response to oxidative stress
- PMID: 16835452
- PMCID: PMC1489289
- DOI: 10.1128/EC.00071-06
TFIIA plays a role in the response to oxidative stress
Abstract
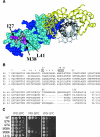
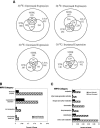
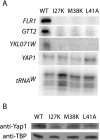
To characterize the role of the general transcription factor TFIIA in the regulation of gene expression by RNA polymerase II, we examined the transcriptional profiles of TFIIA mutants of Saccharomyces cerevisiae using DNA microarrays. Whole-genome expression profiles were determined for three different mutants with mutations in the gene coding for the small subunit of TFIIA, TOA2. Depending on the particular mutant strain, approximately 11 to 27% of the expressed genes exhibit altered message levels. A search for common motifs in the upstream regions of the pool of genes decreased in all three mutants yielded the binding site for Yap1, the transcription factor that regulates the response to oxidative stress. Consistent with a TFIIA-Yap1 connection, the TFIIA mutants are unable to grow under conditions that require the oxidative stress response. Underexpression of Yap1-regulated genes in the TFIIA mutant strains is not the result of decreased expression of Yap1 protein, since immunoblot analysis indicates similar amounts of Yap1 in the wild-type and mutant strains. In addition, intracellular localization studies indicate that both the wild-type and mutant strains localize Yap1 indistinguishably in response to oxidative stress. As such, the decrease in transcription of Yap1-dependent genes in the TFIIA mutant strains appears to reflect a compromised interaction between Yap1 and TFIIA. This hypothesis is supported by the observations that Yap1 and TFIIA interact both in vivo and in vitro. Taken together, these studies demonstrate a dependence of Yap1 on TFIIA function and highlight a new role for TFIIA in the cellular mechanism of defense against reactive oxygen species.
Figures



Similar articles
-
Association of transcription factor IIA with TATA binding protein is required for transcriptional activation of a subset of promoters and cell cycle progression in Saccharomyces cerevisiae.Mol Cell Biol. 1998 May;18(5):2559-70. doi: 10.1128/MCB.18.5.2559. Mol Cell Biol. 1998. PMID: 9566876 Free PMC article.
-
Role for Nhp6, Gcn5, and the Swi/Snf complex in stimulating formation of the TATA-binding protein-TFIIA-DNA complex.Mol Cell Biol. 2004 Sep;24(18):8312-21. doi: 10.1128/MCB.24.18.8312-8321.2004. Mol Cell Biol. 2004. PMID: 15340090 Free PMC article.
-
The yeast FACT complex has a role in transcriptional initiation.Mol Cell Biol. 2005 Jul;25(14):5812-22. doi: 10.1128/MCB.25.14.5812-5822.2005. Mol Cell Biol. 2005. PMID: 15987999 Free PMC article.
-
A facelift for the general transcription factor TFIIA.Biochim Biophys Acta. 2007 Jul-Aug;1769(7-8):429-36. doi: 10.1016/j.bbaexp.2007.04.008. Epub 2007 May 5. Biochim Biophys Acta. 2007. PMID: 17560669 Review.
-
Oxidant-Sensing Pathways in the Responses of Fungal Pathogens to Chemical Stress Signals.Front Microbiol. 2019 Mar 19;10:567. doi: 10.3389/fmicb.2019.00567. eCollection 2019. Front Microbiol. 2019. PMID: 30941117 Free PMC article. Review.
Cited by
-
The Aspergillus fumigatus transcriptional regulator AfYap1 represents the major regulator for defense against reactive oxygen intermediates but is dispensable for pathogenicity in an intranasal mouse infection model.Eukaryot Cell. 2007 Dec;6(12):2290-302. doi: 10.1128/EC.00267-07. Epub 2007 Oct 5. Eukaryot Cell. 2007. PMID: 17921349 Free PMC article.
-
Ino2, activator of yeast phospholipid biosynthetic genes, interacts with basal transcription factors TFIIA and Bdf1.Curr Genet. 2023 Dec;69(4-6):289-300. doi: 10.1007/s00294-023-01277-z. Epub 2023 Nov 10. Curr Genet. 2023. PMID: 37947853 Free PMC article.
-
Transcription activator-like type III effector AvrXa27 depends on OsTFIIAgamma5 for the activation of Xa27 transcription in rice that triggers disease resistance to Xanthomonas oryzae pv. oryzae.Mol Plant Pathol. 2009 Nov;10(6):829-35. doi: 10.1111/j.1364-3703.2009.00567.x. Mol Plant Pathol. 2009. PMID: 19849788 Free PMC article.
-
Reactive Oxygen Species Differentially Modulate the Metabolic and Transcriptomic Response of Endothelial Cells.Antioxidants (Basel). 2022 Feb 21;11(2):434. doi: 10.3390/antiox11020434. Antioxidants (Basel). 2022. PMID: 35204316 Free PMC article.
-
Tracking transcription factor complexes on DNA using total internal reflectance fluorescence protein binding microarrays.Nucleic Acids Res. 2009 Jul;37(13):e94. doi: 10.1093/nar/gkp424. Epub 2009 May 31. Nucleic Acids Res. 2009. PMID: 19487241 Free PMC article.
References
-
- Alarco, A., I. Balan, D. Talibi, N. Mainville, and M. Raymond. 1997. AP1-mediated multidrug resistance in Saccharomyces cerevisiae requires FLR1 encoding a transporter of the major facilitator superfamily. J. Biol. Chem. 272:19304-19313. - PubMed
-
- Athar, M. 2002. Oxidative stress and experimental carcinogensis. Indian J. Exp. Biol. 40:656-667. - PubMed
-
- Auble, D. T., K. E. Hansen, C. G. F. Mueller, W. S. Lane, J. Thorner, and S. Hahn. 1994. Mot1, a global repressor of RNA polymerase II transcription, inhibits TBP binding to DNA by an ATP-dependent mechanism. Genes Dev. 8:1920-1934. - PubMed
-
- Azevedo, D., F. Tacnet, A. Delaunay, C. Rodrigues-Pousada, and M. B. Toledano. 2003. Two redox centers within Yap1 for H2O2 and thiol-reactive chemicals signaling. Free Radic. Biol. Med. 35:889-900. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

