High rate of chimeric gene origination by retroposition in plant genomes
- PMID: 16829590
- PMCID: PMC1533979
- DOI: 10.1105/tpc.106.041905
High rate of chimeric gene origination by retroposition in plant genomes
Abstract
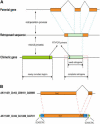
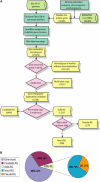
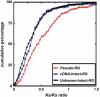
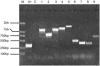
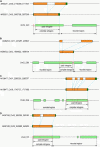
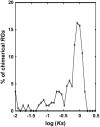
Retroposition is widely found to play essential roles in origination of new mammalian and other animal genes. However, the scarcity of retrogenes in plants has led to the assumption that plant genomes rarely evolve new gene duplicates by retroposition, despite abundant retrotransposons in plants and a reported long terminal repeat (LTR) retrotransposon-mediated mechanism of retroposing cellular genes in maize (Zea mays). We show extensive retropositions in the rice (Oryza sativa) genome, with 1235 identified primary retrogenes. We identified 27 of these primary retrogenes within LTR retrotransposons, confirming a previously observed role of retroelements in generating plant retrogenes. Substitution analyses revealed that the vast majority are subject to negative selection, suggesting, along with expression data and evidence of age, that they are likely functional retrogenes. In addition, 42% of these retrosequences have recruited new exons from flanking regions, generating a large number of chimerical genes. We also identified young chimerical genes, suggesting that gene origination through retroposition is ongoing, with a rate an order of magnitude higher than the rate in primates. Finally, we observed that retropositions have followed an unexpected spatial pattern in which functional retrogenes avoid centromeric regions, while retropseudogenes are randomly distributed. These observations suggest that retroposition is an important mechanism that governs gene evolution in rice and other grass species.
Figures

Similar articles
-
Evolutionary patterns of chimeric retrogenes in Oryza species.Sci Rep. 2019 Nov 27;9(1):17733. doi: 10.1038/s41598-019-54085-2. Sci Rep. 2019. PMID: 31776387 Free PMC article.
-
Genome-wide survey and comparative analysis of LTR retrotransposons and their captured genes in rice and sorghum.PLoS One. 2013 Jul 29;8(7):e71118. doi: 10.1371/journal.pone.0071118. Print 2013. PLoS One. 2013. PMID: 23923055 Free PMC article.
-
The rapid generation of chimerical genes expanding protein diversity in zebrafish.BMC Genomics. 2010 Nov 24;11:657. doi: 10.1186/1471-2164-11-657. BMC Genomics. 2010. PMID: 21106061 Free PMC article.
-
L1 elements, processed pseudogenes and retrogenes in mammalian genomes.IUBMB Life. 2006 Dec;58(12):677-85. doi: 10.1080/15216540601034856. IUBMB Life. 2006. PMID: 17424906 Review.
-
Cancer, Retrogenes, and Evolution.Life (Basel). 2021 Jan 19;11(1):72. doi: 10.3390/life11010072. Life (Basel). 2021. PMID: 33478113 Free PMC article. Review.
Cited by
-
MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity.Nucleic Acids Res. 2012 Apr;40(7):e49. doi: 10.1093/nar/gkr1293. Epub 2012 Jan 4. Nucleic Acids Res. 2012. PMID: 22217600 Free PMC article.
-
A new family of Ty1-copia-like retrotransposons originated in the tomato genome by a recent horizontal transfer event.Genetics. 2009 Apr;181(4):1183-93. doi: 10.1534/genetics.108.099150. Epub 2009 Jan 19. Genetics. 2009. PMID: 19153256 Free PMC article.
-
Genomic localization of AtRE1 and AtRE2, copia-type retrotransposons, in natural variants of Arabidopsis thaliana.Mol Genet Genomics. 2014 Oct;289(5):821-35. doi: 10.1007/s00438-014-0855-z. Epub 2014 Apr 27. Mol Genet Genomics. 2014. PMID: 24770782
-
Small-scale gene duplications played a major role in the recent evolution of wheat chromosome 3B.Genome Biol. 2015 Sep 9;16(1):188. doi: 10.1186/s13059-015-0754-6. Genome Biol. 2015. PMID: 26353816 Free PMC article.
-
The SLEEPER genes: a transposase-derived angiosperm-specific gene family.BMC Plant Biol. 2012 Oct 16;12:192. doi: 10.1186/1471-2229-12-192. BMC Plant Biol. 2012. PMID: 23067104 Free PMC article.
References
-
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815. - PubMed
-
- Arhondakis, S., Auletta, F., Torelli, G., and D'Onofrio, G. (2004). Base composition and expression level of human genes. Gene 325 165–169. - PubMed
-
- Barbazuk, W.B., Bedell, J.A., and Rabinowicz, P.D. (2005). Reduced representation sequencing: A success in maize and a promise for other plant genomes. Bioessays 27 839–848. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

