RNA recognition and cleavage by the SARS coronavirus endoribonuclease
- PMID: 16828802
- PMCID: PMC7118729
- DOI: 10.1016/j.jmb.2006.06.021
RNA recognition and cleavage by the SARS coronavirus endoribonuclease
Abstract
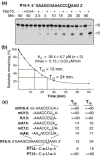
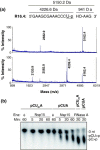
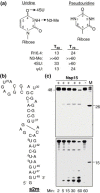
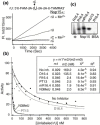
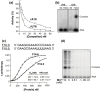
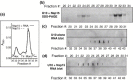
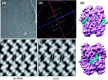
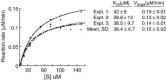
The emerging disease SARS is caused by a novel coronavirus that encodes several unusual RNA-processing enzymes, including non-structural protein 15 (Nsp15), a hexameric endoribonuclease that preferentially cleaves at uridine residues. How Nsp15 recognizes and cleaves RNA is not well understood and is the subject of this study. Based on the analysis of RNA products separated by denaturing gel electrophoresis, Nsp15 has been reported to cleave both 5' and 3' of the uridine. We used several RNAs, including some with nucleotide analogs, and mass spectrometry to determine that Nsp15 cleaves only 3' of the recognition uridylate, with some cleavage 3' of cytidylate. A highly conserved RNA structure in the 3' non-translated region of the SARS virus was cleaved preferentially at one of the unpaired uridylate bases, demonstrating that both RNA structure and base-pairing can affect cleavage by Nsp15. Several modified RNAs that are not cleaved by Nsp15 can bind Nsp15 as competitive inhibitors. The RNA binding affinity of Nsp15 increased with the content of uridylate in substrate RNA and the co-factor Mn(2+). The hexameric form of Nsp15 was found to bind RNA in solution. A two-dimensional crystal of Nsp15 in complex with RNA showed that at least two RNA molecules could be bound per hexamer. Furthermore, an 8.3 A structure of Nsp15 was developed using cyroelectron microscopy, allowing us to generate a model of the Nsp15-RNA complex.
Figures




Similar articles
-
Structural and functional analyses of the severe acute respiratory syndrome coronavirus endoribonuclease Nsp15.J Biol Chem. 2008 Feb 8;283(6):3655-3664. doi: 10.1074/jbc.M708375200. Epub 2007 Nov 28. J Biol Chem. 2008. PMID: 18045871 Free PMC article.
-
Mutational analysis of the SARS virus Nsp15 endoribonuclease: identification of residues affecting hexamer formation.J Mol Biol. 2005 Nov 11;353(5):1106-17. doi: 10.1016/j.jmb.2005.09.007. Epub 2005 Oct 3. J Mol Biol. 2005. PMID: 16216269 Free PMC article.
-
The severe acute respiratory syndrome coronavirus Nsp15 protein is an endoribonuclease that prefers manganese as a cofactor.J Virol. 2004 Nov;78(22):12218-24. doi: 10.1128/JVI.78.22.12218-12224.2004. J Virol. 2004. PMID: 15507608 Free PMC article.
-
The coronavirus nsp15 endoribonuclease: A puzzling protein and pertinent antiviral drug target.Antiviral Res. 2024 Aug;228:105921. doi: 10.1016/j.antiviral.2024.105921. Epub 2024 May 31. Antiviral Res. 2024. PMID: 38825019 Review.
-
An "Old" protein with a new story: Coronavirus endoribonuclease is important for evading host antiviral defenses.Virology. 2018 Apr;517:157-163. doi: 10.1016/j.virol.2017.12.024. Epub 2018 Jan 4. Virology. 2018. PMID: 29307596 Free PMC article. Review.
Cited by
-
A review of SARS-CoV-2 variants and vaccines: Viral properties, mutations, vaccine efficacy, and safety.Infect Med (Beijing). 2023 Sep 12;2(4):247-261. doi: 10.1016/j.imj.2023.08.005. eCollection 2023 Dec. Infect Med (Beijing). 2023. PMID: 38205179 Free PMC article. Review.
-
Atlas of coronavirus replicase structure.Virus Res. 2014 Dec 19;194:49-66. doi: 10.1016/j.virusres.2013.12.004. Epub 2013 Dec 16. Virus Res. 2014. PMID: 24355834 Free PMC article. Review.
-
Essential Oils as Antiviral Agents. Potential of Essential Oils to Treat SARS-CoV-2 Infection: An In-Silico Investigation.Int J Mol Sci. 2020 May 12;21(10):3426. doi: 10.3390/ijms21103426. Int J Mol Sci. 2020. PMID: 32408699 Free PMC article.
-
Feasibility of Known RNA Polymerase Inhibitors as Anti-SARS-CoV-2 Drugs.Pathogens. 2020 Apr 26;9(5):320. doi: 10.3390/pathogens9050320. Pathogens. 2020. PMID: 32357471 Free PMC article.
-
Cryo-EM structures of the SARS-CoV-2 endoribonuclease Nsp15 reveal insight into nuclease specificity and dynamics.Nat Commun. 2021 Jan 27;12(1):636. doi: 10.1038/s41467-020-20608-z. Nat Commun. 2021. PMID: 33504779 Free PMC article.
References
-
- Holmes K.V., Enjuanes L. Virology. The SARS coronavirus: a postgenomic era. Science. 2003;300:1377–1378. - PubMed
-
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

