Cell-culture assays reveal the importance of retroviral vector design for insertional genotoxicity
- PMID: 16825499
- PMCID: PMC1895590
- DOI: 10.1182/blood-2005-08-024976
Cell-culture assays reveal the importance of retroviral vector design for insertional genotoxicity
Abstract
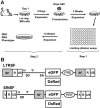
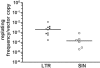
Retroviral vectors with long terminal repeats (LTRs), which contain strong enhancer/promoter sequences at both ends of their genome, are widely used for stable gene transfer into hematopoietic cells. However, recent clinical data and mouse models point to insertional activation of cellular proto-oncogenes as a dose-limiting side effect of retroviral gene delivery that potentially induces leukemia. Self-inactivating (SIN) retroviral vectors do not contain the terminal repetition of the enhancer/promoter, theoretically attenuating the interaction with neighboring cellular genes. With a new assay based on in vitro expansion of primary murine hematopoietic cells and selection in limiting dilution, we showed that SIN vectors using a strong internal retroviral enhancer/promoter may also transform cells by insertional mutagenesis. Most transformed clones, including those obtained after dose escalation of SIN vectors, showed insertions upstream of the third exon of Evi1 and in reverse orientation to its transcriptional orientation. Normalizing for the vector copy number, we found the transforming capacity of SIN vectors to be significantly reduced when compared with corresponding LTR vectors. Additional modifications of SIN vectors may further increase safety. Improved cell-culture assays will likely play an important role in the evaluation of insertional mutagenesis.
Figures

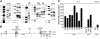

Similar articles
-
Self-inactivating retroviral vector-mediated gene transfer induces oncogene activation and immortalization of primary murine bone marrow cells.Mol Ther. 2009 Nov;17(11):1910-8. doi: 10.1038/mt.2009.172. Epub 2009 Jul 28. Mol Ther. 2009. PMID: 19638958 Free PMC article.
-
Physiological promoters reduce the genotoxic risk of integrating gene vectors.Mol Ther. 2008 Apr;16(4):718-25. doi: 10.1038/mt.2008.5. Epub 2008 Mar 4. Mol Ther. 2008. PMID: 18334985
-
Leukemia induction after a single retroviral vector insertion in Evi1 or Prdm16.Leukemia. 2008 Aug;22(8):1519-28. doi: 10.1038/leu.2008.118. Epub 2008 May 22. Leukemia. 2008. PMID: 18496560
-
Retroviral Insertional Mutagenesis in Humans: Evidence for Four Genetic Mechanisms Promoting Expansion of Cell Clones.Mol Ther. 2020 Feb 5;28(2):352-356. doi: 10.1016/j.ymthe.2019.12.009. Epub 2020 Jan 7. Mol Ther. 2020. PMID: 31951833 Free PMC article. Review.
-
Viral Vectors: The Road to Reducing Genotoxicity.Toxicol Sci. 2017 Feb;155(2):315-325. doi: 10.1093/toxsci/kfw220. Epub 2016 Nov 1. Toxicol Sci. 2017. PMID: 27803388 Review.
Cited by
-
Evaluation of γ-retroviral vectors that mediate the inducible expression of IL-12 for clinical application.J Immunother. 2012 Jun;35(5):430-9. doi: 10.1097/CJI.0b013e31825898e8. J Immunother. 2012. PMID: 22576348 Free PMC article.
-
Pharmacological targeting of the thrombomodulin-activated protein C pathway mitigates radiation toxicity.Nat Med. 2012 Jul;18(7):1123-9. doi: 10.1038/nm.2813. Nat Med. 2012. PMID: 22729286 Free PMC article.
-
Hematopoietic Stem Cell Gene Therapy for Cystinosis: From Bench-to-Bedside.Cells. 2021 Nov 23;10(12):3273. doi: 10.3390/cells10123273. Cells. 2021. PMID: 34943781 Free PMC article. Review.
-
Preventing and exploiting the oncogenic potential of integrating gene vectors.J Clin Invest. 2009 Apr;119(4):755-8. doi: 10.1172/jci38831. J Clin Invest. 2009. PMID: 19348042 Free PMC article.
-
Scale-up and manufacturing of clinical-grade self-inactivating γ-retroviral vectors by transient transfection.Gene Ther. 2012 Mar;19(3):246-54. doi: 10.1038/gt.2011.102. Epub 2011 Jul 14. Gene Ther. 2012. PMID: 21753795 Free PMC article.
References
-
- Hacein-Bey-Abina S, Le Deist F, Carlier F, et al. Sustained correction of X-linked severe combined immunodeficiency by ex vivo gene therapy. N Engl J Med. 2002;346: 1185-1193. - PubMed
-
- Aiuti A, Slavin S, Aker M, et al. Correction of ADA-SCID by stem cell gene therapy combined with nonmyeloablative conditioning. Science. 2002; 296: 2410-2413. - PubMed
-
- Gaspar HB, Parsley KL, Howe S, et al. Gene therapy of X-linked severe combined immunodeficiency by use of a pseudotyped gammaretroviral vector. Lancet. 2004;364: 2181-2187. - PubMed
-
- Ott MG, Schmidt M, Schwarzwaelder K, et al. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat Med. 2006;12: 401-409. - PubMed
-
- Thomas CE, Ehrhardt A, Kay MA. Progress and problems with the use of viral vectors for gene therapy. Nat Rev Genet. 2003;4: 346-358. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

