Heparan sulphation patterns generated by specific heparan sulfotransferase enzymes direct distinct aspects of retinal axon guidance at the optic chiasm
- PMID: 16807321
- PMCID: PMC6673919
- DOI: 10.1523/JNEUROSCI.0505-06.2006
Heparan sulphation patterns generated by specific heparan sulfotransferase enzymes direct distinct aspects of retinal axon guidance at the optic chiasm
Abstract
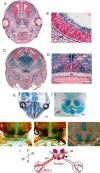
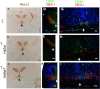
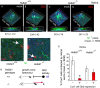
Retinal ganglion cell (RGC) axons from each eye execute a series of maneuvers as they converge on the ventral surface of the brain at the optic chiasm for sorting into the optic tracts. Heparan sulfate proteoglycans (HSPGs) are extracellular glycoproteins involved in cell-surface interactions. HSPGs exhibit massive structural diversity, conferred partly by extensive post-translational modification including differential sulfation. Here we examine the roles of HSPG sulfation in RGC axon guidance at the chiasm. We identified different axon navigation phenotypes in two heparan sulfate sulfotransferase (Hst) mutant embryos, Hs2st-/- and Hs6st1-/-, each lacking an enzyme that catalyzes a particular HSPG modification. Hs2st-/- embryos display axon disorganization at the chiasm. Hs6st1-/- embryos exhibit prolific inter-retinal innervation. We show that RGCs express Hs2st and Hs6st1 and that navigation errors made by their axons coincide with regions of high Hs2st and/or Hs6st1 expression at the chiasm. Slit proteins are expressed at particular locations in the retina and around the chiasm and are normally deployed to prevent axons entering inappropriate territories. We show that Hs2st and/or Hs6st1 expression coincides with Slit expression domains at locations where RGC axons make navigation errors in Hs2st-/- and Hs6st1-/- mutants and that Hs6st1-/- RGC axons are less sensitive to Slit2 repulsion than their wild-type counterparts in vitro. We suggest that (1) Hs2st and Hs6st1 are each deployed to generate distinct patterns of heparan sulfation on RGCs and at the optic chiasm and (2) this differential sulfation directs retinal axons through the chiasm, at least in part by modulating the response of the navigating growth cone to Slit proteins.
Figures


Similar articles
-
Heparan Sulfate Sulfation by Hs2st Restricts Astroglial Precursor Somal Translocation in Developing Mouse Forebrain by a Non-Cell-Autonomous Mechanism.J Neurosci. 2019 Feb 20;39(8):1386-1404. doi: 10.1523/JNEUROSCI.1747-17.2018. Epub 2019 Jan 7. J Neurosci. 2019. PMID: 30617207 Free PMC article.
-
Analysis of axon guidance defects at the optic chiasm in heparan sulphate sulphotransferase compound mutant mice.J Anat. 2011 Dec;219(6):734-42. doi: 10.1111/j.1469-7580.2011.01432.x. Epub 2011 Sep 26. J Anat. 2011. PMID: 21951307 Free PMC article.
-
Heparan sulfate sugar modifications mediate the functions of slits and other factors needed for mouse forebrain commissure development.J Neurosci. 2011 Feb 9;31(6):1955-70. doi: 10.1523/JNEUROSCI.2579-10.2011. J Neurosci. 2011. PMID: 21307234 Free PMC article.
-
[Exploration of the molecular mechanism of ocular development and the creation of animal models for ocular diseases].Nippon Ganka Gakkai Zasshi. 2010 Mar;114(3):280-96; discussion 297. Nippon Ganka Gakkai Zasshi. 2010. PMID: 20387539 Review. Japanese.
-
Retinal axon growth at the optic chiasm: to cross or not to cross.Annu Rev Neurosci. 2008;31:295-315. doi: 10.1146/annurev.neuro.31.060407.125609. Annu Rev Neurosci. 2008. PMID: 18558857 Review.
Cited by
-
2- and 6-O-sulfated proteoglycans have distinct and complementary roles in cranial axon guidance and motor neuron migration.Development. 2016 Jun 1;143(11):1907-13. doi: 10.1242/dev.126854. Epub 2016 Apr 5. Development. 2016. PMID: 27048738 Free PMC article.
-
Heparan sulfate 6-O-sulfotransferase isoform-dependent regulatory effects of heparin on the activities of various proteases in mast cells and the biosynthesis of 6-O-sulfated heparin.J Biol Chem. 2013 Feb 8;288(6):3705-17. doi: 10.1074/jbc.M112.416651. Epub 2012 Dec 6. J Biol Chem. 2013. PMID: 23223449 Free PMC article.
-
A molecular mechanism for the heparan sulfate dependence of slit-robo signaling.J Biol Chem. 2006 Dec 22;281(51):39693-8. doi: 10.1074/jbc.M609384200. Epub 2006 Oct 24. J Biol Chem. 2006. PMID: 17062560 Free PMC article.
-
Heparan Sulfate Sulfation by Hs2st Restricts Astroglial Precursor Somal Translocation in Developing Mouse Forebrain by a Non-Cell-Autonomous Mechanism.J Neurosci. 2019 Feb 20;39(8):1386-1404. doi: 10.1523/JNEUROSCI.1747-17.2018. Epub 2019 Jan 7. J Neurosci. 2019. PMID: 30617207 Free PMC article.
-
Visualizing mechanosensory endings of TrkC-expressing neurons in HS3ST-2-hPLAP mice.J Comp Neurol. 2008 Dec 1;511(4):543-56. doi: 10.1002/cne.21862. J Comp Neurol. 2008. PMID: 18839409 Free PMC article.
References
-
- Bulow HE, Hobert O (2004). Differential sulphations and epimerisation define heparan sulphate specificity in nervous system development. Neuron 41:723–736. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
