SUMO: regulating the regulator
- PMID: 16805918
- PMCID: PMC1524954
- DOI: 10.1186/1747-1028-1-13
SUMO: regulating the regulator
Abstract
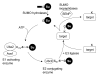
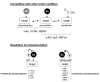
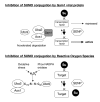
Post-translational modifiers of the SUMO (Small Ubiquitin-related Modifier) family have emerged as key regulators of protein function and fate. While the past few years have seen an enormous increase in knowledge on SUMO enzymes, substrates, and consequences of modification, regulation of SUMO conjugation is far from being understood. This brief review will provide an overview on recent advances concerning (i) the interplay between sumoylation and other post-translational modifications at the level of individual targets and (ii) global regulation of SUMO conjugation and deconjugation.
Figures
Similar articles
-
SUMO under stress.Biochem Soc Trans. 2008 Oct;36(Pt 5):874-8. doi: 10.1042/BST0360874. Biochem Soc Trans. 2008. PMID: 18793154 Review.
-
[Advances on SUMO substrates in Arabidopsis].Yi Chuan. 2013 Jun;35(6):727-34. doi: 10.3724/sp.j.1005.2013.00727. Yi Chuan. 2013. PMID: 23774017 Review. Chinese.
-
Molecular mechanisms in SUMO conjugation.Biochem Soc Trans. 2020 Feb 28;48(1):123-135. doi: 10.1042/BST20190357. Biochem Soc Trans. 2020. PMID: 31872228 Review.
-
SUMO: getting it on.Biochem Soc Trans. 2007 Dec;35(Pt 6):1409-13. doi: 10.1042/BST0351409. Biochem Soc Trans. 2007. PMID: 18031233 Review.
-
Sumoylation, a post-translational regulatory process in plants.Curr Opin Plant Biol. 2007 Oct;10(5):495-502. doi: 10.1016/j.pbi.2007.07.002. Epub 2007 Aug 27. Curr Opin Plant Biol. 2007. PMID: 17720613 Review.
Cited by
-
SUMO: a (oxidative) stressed protein.Neuromolecular Med. 2013 Dec;15(4):707-19. doi: 10.1007/s12017-013-8266-6. Epub 2013 Sep 20. Neuromolecular Med. 2013. PMID: 24052421 Review.
-
Interaction between geminivirus replication protein and the SUMO-conjugating enzyme is required for viral infection.J Virol. 2011 Oct;85(19):9789-800. doi: 10.1128/JVI.02566-10. Epub 2011 Jul 20. J Virol. 2011. PMID: 21775461 Free PMC article.
-
Enzymatic regulation and functional relevance of NOX5.Curr Pharm Des. 2015;21(41):5999-6008. doi: 10.2174/1381612821666151029111528. Curr Pharm Des. 2015. PMID: 26510438 Free PMC article. Review.
-
Regulation of cold signaling by sumoylation of ICE1.Plant Signal Behav. 2008 Jan;3(1):52-3. doi: 10.4161/psb.3.1.4865. Plant Signal Behav. 2008. PMID: 19704769 Free PMC article.
-
Sumoylation of amyloid precursor protein negatively regulates Abeta aggregate levels.Biochem Biophys Res Commun. 2008 Oct 3;374(4):673-8. doi: 10.1016/j.bbrc.2008.07.109. Epub 2008 Jul 31. Biochem Biophys Res Commun. 2008. PMID: 18675254 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

