Critical roles of the immunoglobulin intronic enhancers in maintaining the sequential rearrangement of IgH and Igk loci
- PMID: 16785310
- PMCID: PMC2118354
- DOI: 10.1084/jem.20052310
Critical roles of the immunoglobulin intronic enhancers in maintaining the sequential rearrangement of IgH and Igk loci
Abstract
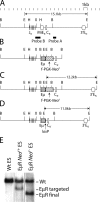
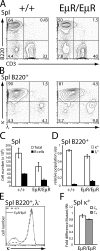
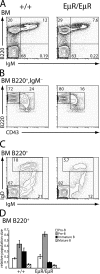
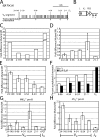
V(D)J recombination of immunoglobulin (Ig) heavy (IgH) and light chain genes occurs sequentially in the pro- and pre-B cells. To identify cis-elements that dictate this order of rearrangement, we replaced the endogenous matrix attachment region/Igk intronic enhancer (MiE(kappa)) with its heavy chain counterpart (Emu) in mice. This replacement, denoted EmuR, substantially increases the accessibility of both V(kappa) and J(kappa) loci to V(D)J recombinase in pro-B cells and induces Igk rearrangement in these cells. However, EmuR does not support Igk rearrangement in pre-B cells. Similar to that in MiE(kappa)(-/-) pre-B cells, the accessibility of V(kappa) segments to V(D)J recombinase is considerably reduced in EmuR pre-B cells when compared with wild-type pre-B cells. Therefore, Emu and MiE(kappa) play developmental stage-specific roles in maintaining the sequential rearrangement of IgH and Igk loci by promoting the accessibility of V, D, and J loci to the V(D)J recombinase.
Figures



Similar articles
-
Immunoglobulin light chain gene rearrangements display hierarchy in absence of selection for functionality in precursor-B-ALL.Leukemia. 2002 Aug;16(8):1448-53. doi: 10.1038/sj.leu.2402548. Leukemia. 2002. PMID: 12145684
-
Chromatin remodeling at the Ig loci prior to V(D)J recombination.J Immunol. 2001 Jul 15;167(2):866-74. doi: 10.4049/jimmunol.167.2.866. J Immunol. 2001. PMID: 11441093
-
Ig heavy chain protein controls B cell development by regulating germ-line transcription and retargeting V(D)J recombination.J Immunol. 1994 Aug 15;153(4):1645-57. J Immunol. 1994. PMID: 8046237
-
Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus.Annu Rev Immunol. 2006;24:541-70. doi: 10.1146/annurev.immunol.23.021704.115830. Annu Rev Immunol. 2006. PMID: 16551259 Review.
-
Regulation of activation and recombination of the murine Igkappa locus.Immunol Rev. 2004 Aug;200:215-23. doi: 10.1111/j.0105-2896.2004.00157.x. Immunol Rev. 2004. PMID: 15242407 Review.
Cited by
-
A developmentally controlled competitive STAT5-PU.1 DNA binding mechanism regulates activity of the Ig κ E3' enhancer.J Immunol. 2012 Mar 1;188(5):2276-84. doi: 10.4049/jimmunol.1102239. Epub 2012 Jan 25. J Immunol. 2012. PMID: 22279106 Free PMC article.
-
Identification of multi-loci hubs from 4C-seq demonstrates the functional importance of simultaneous interactions.Nucleic Acids Res. 2016 Oct 14;44(18):8714-8725. doi: 10.1093/nar/gkw568. Epub 2016 Jul 20. Nucleic Acids Res. 2016. PMID: 27439714 Free PMC article.
-
Anti-aging Effect of Transplanted Amniotic Membrane Mesenchymal Stem Cells in a Premature Aging Model of Bmi-1 Deficiency.Sci Rep. 2015 Sep 15;5:13975. doi: 10.1038/srep13975. Sci Rep. 2015. Retraction in: Sci Rep. 2024 Nov 26;14(1):29315. doi: 10.1038/s41598-024-80756-w. PMID: 26370922 Free PMC article. Retracted.
-
Pre-B cell receptor signaling induces immunoglobulin κ locus accessibility by functional redistribution of enhancer-mediated chromatin interactions.PLoS Biol. 2014 Feb 18;12(2):e1001791. doi: 10.1371/journal.pbio.1001791. eCollection 2014 Feb. PLoS Biol. 2014. PMID: 24558349 Free PMC article.
-
Phospholipase Cgamma2 contributes to light-chain gene activation and receptor editing.Mol Cell Biol. 2007 Sep;27(17):5957-67. doi: 10.1128/MCB.02273-06. Epub 2007 Jun 25. Mol Cell Biol. 2007. PMID: 17591700 Free PMC article.
References
-
- Bassing, C.H., W. Swat, and F.W. Alt. 2002. The mechanism and regulation of chromosomal V(D)J recombination. Cell. 109:S45–S55. - PubMed
-
- Yancopoulos, G.D., and F.W. Alt. 1985. Developmentally controlled and tissue-specific expression of unrearranged Vh gene segments. Cell. 40:271–281. - PubMed
-
- Alt, F.W., T.K. Blackwell, and G.D. Yancopoulos. 1987. Development of the primary antibody repertoire. Science. 238:1079–1087. - PubMed
-
- Stanhope-Baker, P., K.M. Hudson, A.L. Shaffer, A. Constantinescu, and M.S. Schlissel. 1996. Cell type-specific chromatin structure determines the targeting of V(D)J recombinase activity in vitro. Cell. 85:887–897. - PubMed
-
- Sleckman, B.P., J.R. Gorman, and F.W. Alt. 1996. Accessibility control of antigen-receptor variable-region gene assembly: role of cis-acting elements. Annu. Rev. Immunol. 14:459–481. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

