Interaction between PCNA and diubiquitinated Mcm10 is essential for cell growth in budding yeast
- PMID: 16782870
- PMCID: PMC1489165
- DOI: 10.1128/MCB.02062-05
Interaction between PCNA and diubiquitinated Mcm10 is essential for cell growth in budding yeast
Abstract
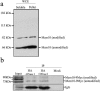
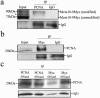
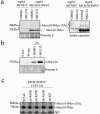
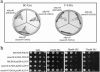
The minichromosome maintenance protein 10 (Mcm10) is an evolutionarily conserved factor that is essential for replication initiation and elongation. Mcm10 is part of the eukaryotic replication fork and interacts with a variety of proteins, including the Mcm2-7 helicase and DNA polymerase alpha/primase complexes. A motif search revealed a match to the proliferating cell nuclear antigen (PCNA)-interacting protein (PIP) box in Mcm10. Here, we demonstrate a direct interaction between Mcm10 and PCNA that is alleviated by mutations in conserved residues of the PIP box. Interestingly, only the diubiquitinated form of Mcm10 binds to PCNA. Diubiquitination of Mcm10 is cell cycle regulated; it first appears in late G(1) and persists throughout S phase. During this time, diubiquitinated Mcm10 is associated with chromatin, suggesting a direct role in DNA replication. Surprisingly, a Y245A substitution in the PIP box of Mcm10 that inhibits the interaction with PCNA abolishes cell proliferation. This severe-growth phenotype, which has not been observed for analogous mutations in other PCNA-interacting proteins, is rescued by a compensatory mutation in PCNA that restores interaction with Mcm10-Y245A. Taken together, our results suggest that diubiquitinated Mcm10 interacts with PCNA to facilitate an essential step in DNA elongation.
Figures
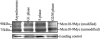

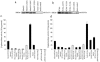
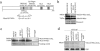
Similar articles
-
Mcm10 and the MCM2-7 complex interact to initiate DNA synthesis and to release replication factors from origins.Genes Dev. 2000 Apr 15;14(8):913-26. Genes Dev. 2000. PMID: 10783164 Free PMC article.
-
Mcm10 regulates the stability and chromatin association of DNA polymerase-alpha.Mol Cell. 2004 Oct 22;16(2):173-85. doi: 10.1016/j.molcel.2004.09.017. Mol Cell. 2004. PMID: 15494305
-
An intact Mcm10 coiled-coil interaction surface is important for origin melting, helicase assembly and the recruitment of Pol-α to Mcm2-7.Nucleic Acids Res. 2017 Jul 7;45(12):7261-7275. doi: 10.1093/nar/gkx438. Nucleic Acids Res. 2017. PMID: 28510759 Free PMC article.
-
Enigmatic roles of Mcm10 in DNA replication.Trends Biochem Sci. 2013 Apr;38(4):184-94. doi: 10.1016/j.tibs.2012.12.003. Epub 2013 Jan 17. Trends Biochem Sci. 2013. PMID: 23332289 Free PMC article. Review.
-
Mcm10: A Dynamic Scaffold at Eukaryotic Replication Forks.Genes (Basel). 2017 Feb 17;8(2):73. doi: 10.3390/genes8020073. Genes (Basel). 2017. PMID: 28218679 Free PMC article. Review.
Cited by
-
Four decades of Eukaryotic DNA replication: From yeast genetics to high-resolution cryo-EM structures of the replisome.Proc Natl Acad Sci U S A. 2024 Oct 15;121(42):e2415231121. doi: 10.1073/pnas.2415231121. Epub 2024 Oct 4. Proc Natl Acad Sci U S A. 2024. PMID: 39365830
-
RRM3 regulates epigenetic conversions in Saccharomyces cerevisiae in conjunction with Chromatin Assembly Factor I.Nucleus. 2016 Jul 3;7(4):405-14. doi: 10.1080/19491034.2016.1212796. Nucleus. 2016. PMID: 27645054 Free PMC article.
-
Control of DNA Replication Initiation by Ubiquitin.Cells. 2018 Sep 20;7(10):146. doi: 10.3390/cells7100146. Cells. 2018. PMID: 30241373 Free PMC article. Review.
-
CDC28 phosphorylates Cac1p and regulates the association of chromatin assembly factor I with chromatin.Cell Cycle. 2015;14(1):74-85. doi: 10.4161/15384101.2014.973745. Cell Cycle. 2015. PMID: 25602519 Free PMC article.
-
The anti-parasitic agent suramin and several of its analogues are inhibitors of the DNA binding protein Mcm10.Open Biol. 2019 Aug 30;9(8):190117. doi: 10.1098/rsob.190117. Epub 2019 Aug 14. Open Biol. 2019. PMID: 31409229 Free PMC article.
References
-
- Bell, S. P., and A. Dutta. 2002. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71:333-374. - PubMed
-
- Bielinsky, A. K., and S. A. Gerbi. 1999. Chromosomal ARS1 has a single leading strand start site. Mol. Cell 3:477-486. - PubMed
-
- Bielinsky, A. K., and S. A. Gerbi. 1998. Discrete start sites for DNA synthesis in the yeast ARS1 origin. Science 279:95-98. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
