The conserved AAUAAA hexamer of the poly(A) signal can act alone to trigger a stable decrease in RNA polymerase II transcription velocity
- PMID: 16775304
- PMCID: PMC1524889
- DOI: 10.1261/rna.103206
The conserved AAUAAA hexamer of the poly(A) signal can act alone to trigger a stable decrease in RNA polymerase II transcription velocity
Abstract
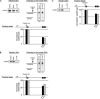
In vivo the poly(A) signal not only directs 3'-end processing but also controls the rate and extent of transcription. Thus, upon crossing the poly(A) signal RNA polymerase II first pauses and then terminates. We show that the G/U-rich region of the poly(A) signal, although required for termination in vivo, is not required for poly(A)-dependent pausing either in vivo or in vitro. Consistent with this, neither CstF, which recognizes the G/U-rich element, nor the polymerase CTD, which binds CstF, is required for pausing. The only part of the poly(A) signal required to direct the polymerase to pause is the AAUAAA hexamer. The effect of the hexamer on the polymerase is long lasting--in many situations polymerases over 1 kb downstream of the hexamer continue to exhibit delayed progress down the template in vivo. The hexamer is the first part of the poly(A) signal to emerge from the polymerase and may play a role independent of the rest of the poly(A) signal in paving the way for subsequent events such as 3'-end processing and termination of transcription.
Figures



Similar articles
-
The poly(A)-dependent transcriptional pause is mediated by CPSF acting on the body of the polymerase.Nat Struct Mol Biol. 2007 Jul;14(7):662-9. doi: 10.1038/nsmb1253. Epub 2007 Jun 17. Nat Struct Mol Biol. 2007. PMID: 17572685
-
3' RNA processing efficiency plays a primary role in generating termination-competent RNA polymerase II elongation complexes.Mol Cell Biol. 1993 Jun;13(6):3472-80. doi: 10.1128/mcb.13.6.3472-3480.1993. Mol Cell Biol. 1993. PMID: 7684499 Free PMC article.
-
A functional mRNA polyadenylation signal is required for transcription termination by RNA polymerase II.Genes Dev. 1988 Apr;2(4):440-52. doi: 10.1101/gad.2.4.440. Genes Dev. 1988. PMID: 2836265
-
Transcription termination and 3' processing: the end is in site!Cell. 1985 Jun;41(2):349-59. doi: 10.1016/s0092-8674(85)80007-6. Cell. 1985. PMID: 2580642 Review. No abstract available.
-
Transcriptional termination in mammals: Stopping the RNA polymerase II juggernaut.Science. 2016 Jun 10;352(6291):aad9926. doi: 10.1126/science.aad9926. Science. 2016. PMID: 27284201 Free PMC article. Review.
Cited by
-
Prolonged α-amanitin treatment of cells for studying mutated polymerases causes degradation of DSIF160 and other proteins.RNA. 2012 Feb;18(2):222-9. doi: 10.1261/rna.030452.111. Epub 2011 Dec 22. RNA. 2012. PMID: 22194310 Free PMC article.
-
The transcriptional cycle of HIV-1 in real-time and live cells.J Cell Biol. 2007 Oct 22;179(2):291-304. doi: 10.1083/jcb.200706018. J Cell Biol. 2007. PMID: 17954611 Free PMC article.
-
Control of RNA Pol II Speed by PNUTS-PP1 and Spt5 Dephosphorylation Facilitates Termination by a "Sitting Duck Torpedo" Mechanism.Mol Cell. 2019 Dec 19;76(6):896-908.e4. doi: 10.1016/j.molcel.2019.09.031. Epub 2019 Oct 30. Mol Cell. 2019. PMID: 31677974 Free PMC article.
-
The Reb1-homologue Ydr026c/Nsi1 is required for efficient RNA polymerase I termination in yeast.EMBO J. 2012 Aug 15;31(16):3480-93. doi: 10.1038/emboj.2012.185. Epub 2012 Jul 17. EMBO J. 2012. PMID: 22805593 Free PMC article.
-
RNA polymerase II pausing downstream of core histone genes is different from genes producing polyadenylated transcripts.PLoS One. 2012;7(6):e38769. doi: 10.1371/journal.pone.0038769. Epub 2012 Jun 11. PLoS One. 2012. PMID: 22701709 Free PMC article.
References
-
- Bird G., Fong N., Gatlin J.C., Farabaugh S., Bentley D.L. Ribozyme cleavage reveals connections between mRNA release from the site of transcription and Pre-mRNA processing. Mol. Cell. 2005;20:747–758. - PubMed
-
- Calvo O., Manley J.L. Strange bedfellows: Polyadenylation factors at the promoter. Genes & Dev. 2003;17:1321–1327. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
