A comprehensive structure-function analysis of Arabidopsis SNI1 defines essential regions and transcriptional repressor activity
- PMID: 16766691
- PMCID: PMC1488919
- DOI: 10.1105/tpc.105.039677
A comprehensive structure-function analysis of Arabidopsis SNI1 defines essential regions and transcriptional repressor activity
Abstract
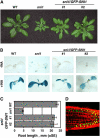
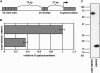
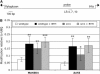
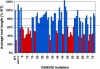
The expression of systemic acquired resistance (SAR) in plants involves the upregulation of many Pathogenesis-Related (PR) genes, which work in concert to confer resistance to a broad spectrum of pathogens. Because SAR is a costly process, SAR-associated transcription must be tightly regulated. Arabidopsis thaliana SNI1 (for Suppressor of NPR1, Inducible) is a negative regulator of SAR required to dampen the basal expression of PR genes. Whole genome transcriptional profiling showed that in the sni1 mutant, Nonexpresser of PR genes (NPR1)-dependent benzothiadiazole S-methylester-responsive genes were specifically derepressed. Interestingly, SNI1 also repressed transcription when expressed in yeast, suggesting that it functions as an active transcriptional repressor through a highly conserved mechanism. Chromatin immunoprecipitation indicated that histone modification may be involved in SNI1-mediated repression. Sequence comparison with orthologs in other plant species and a saturating NAAIRS-scanning mutagenesis of SNI1 identified regions in SNI1 that are required for its activity. The structural similarity of SNI1 to Armadillo repeat proteins implies that SNI1 may form a scaffold for interaction with proteins that modulate transcription.
Figures



Similar articles
-
The Arabidopsis PR-1 promoter contains multiple integration sites for the coactivator NPR1 and the repressor SNI1.Plant Physiol. 2010 Dec;154(4):1805-18. doi: 10.1104/pp.110.165563. Epub 2010 Oct 8. Plant Physiol. 2010. PMID: 20935179 Free PMC article.
-
DNA repair proteins are directly involved in regulation of gene expression during plant immune response.Cell Host Microbe. 2011 Feb 17;9(2):115-24. doi: 10.1016/j.chom.2011.01.011. Cell Host Microbe. 2011. PMID: 21320694
-
Regulation of systemic acquired resistance by NPR1 and its partners.Novartis Found Symp. 2001;236:165-73; discussion 173-5. doi: 10.1002/9780470515778.ch12. Novartis Found Symp. 2001. PMID: 11387978
-
[Structure and function analysis of Arabidopsis thaliana SRO protein family].Yi Chuan. 2013 Oct;35(10):1189-97. doi: 10.3724/sp.j.1005.2013.01189. Yi Chuan. 2013. PMID: 24459892 Review. Chinese.
-
Spatiotemporal expression control correlates with intragenic scaffold matrix attachment regions (S/MARs) in Arabidopsis thaliana.PLoS Comput Biol. 2006 Mar;2(3):e21. doi: 10.1371/journal.pcbi.0020021. Epub 2006 Mar 31. PLoS Comput Biol. 2006. PMID: 16604187 Free PMC article. Review.
Cited by
-
The Arabidopsis PR-1 promoter contains multiple integration sites for the coactivator NPR1 and the repressor SNI1.Plant Physiol. 2010 Dec;154(4):1805-18. doi: 10.1104/pp.110.165563. Epub 2010 Oct 8. Plant Physiol. 2010. PMID: 20935179 Free PMC article.
-
The coactivator function of Arabidopsis NPR1 requires the core of its BTB/POZ domain and the oxidation of C-terminal cysteines.Plant Cell. 2006 Dec;18(12):3670-85. doi: 10.1105/tpc.106.046953. Epub 2006 Dec 15. Plant Cell. 2006. PMID: 17172357 Free PMC article.
-
Negative regulation of systemic acquired resistance by replication factor C subunit3 in Arabidopsis.Plant Physiol. 2009 Aug;150(4):2009-17. doi: 10.1104/pp.109.138321. Epub 2009 May 29. Plant Physiol. 2009. PMID: 19482917 Free PMC article.
-
Heterosis associated gene expression in maize embryos 6 days after fertilization exhibits additive, dominant and overdominant pattern.Plant Mol Biol. 2007 Feb;63(3):381-91. doi: 10.1007/s11103-006-9095-x. Epub 2006 Nov 3. Plant Mol Biol. 2007. PMID: 17082872
-
A comprehensive mutational analysis of the Arabidopsis resistance protein RPW8.2 reveals key amino acids for defense activation and protein targeting.Plant Cell. 2013 Oct;25(10):4242-61. doi: 10.1105/tpc.113.117226. Epub 2013 Oct 22. Plant Cell. 2013. PMID: 24151293 Free PMC article.
References
-
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215 403–410. - PubMed
-
- Axtell, M.J., Chisholm, S.T., Dahlbeck, D., and Staskawicz, B.J. (2003). Genetic and molecular evidence that the Pseudomonas syringae type III effector protein AvrRpt2 is a cysteine protease. Mol. Microbiol. 49 1537–1546. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

