Leukosialin (CD43) defines hematopoietic progenitors in human embryonic stem cell differentiation cultures
- PMID: 16757688
- PMCID: PMC1895535
- DOI: 10.1182/blood-2006-02-003327
Leukosialin (CD43) defines hematopoietic progenitors in human embryonic stem cell differentiation cultures
Abstract
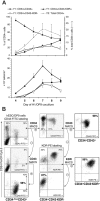
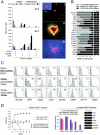
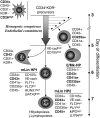
During hematopoietic differentiation of human embryonic stem cells (hESCs), early hematopoietic progenitors arise along with endothelial cells within the CD34(+) population. Although hESC-derived hematopoietic progenitors have been previously identified by functional assays, their phenotype has not been defined. Here, using hESC differentiation in coculture with OP9 stromal cells, we demonstrate that early progenitors committed to hematopoietic development could be identified by surface expression of leukosialin (CD43). CD43 was detected on all types of emerging clonogenic progenitors before expression of CD45, persisted on differentiating hematopoietic cells, and reliably separated the hematopoietic CD34(+) population from CD34(+)CD43(-)CD31(+)KDR(+) endothelial and CD34(+)CD43(-)CD31(-)KDR(-) mesenchymal cells. Furthermore, we demonstrated that the first-appearing CD34(+)CD43(+)CD235a(+)CD41a(+/-)CD45(-) cells represent precommitted erythro-megakaryocytic progenitors. Multipotent lymphohematopoietic progenitors were generated later as CD34(+)CD43(+)CD41a(-)CD235a(-)CD45(-) cells. These cells were negative for lineage-specific markers (Lin(-)), expressed KDR, VE-cadherin, and CD105 endothelial proteins, and expressed GATA-2, GATA-3, RUNX1, C-MYB transcription factors that typify initial stages of definitive hematopoiesis originating from endothelial-like precursors. Acquisition of CD45 expression by CD34(+)CD43(+)CD45(-)Lin(-) cells was associated with progressive myeloid commitment and a decrease of B-lymphoid potential. CD34(+)CD43(+)CD45(+)Lin(-) cells were largely devoid of VE-cadherin and KDR expression and had a distinct FLT3(high)GATA3(low)RUNX1(low)PU1(high)MPO(high)IL7RA(high) gene expression profile.
Figures




Similar articles
-
Hematopoietic and endothelial differentiation of human induced pluripotent stem cells.Stem Cells. 2009 Mar;27(3):559-67. doi: 10.1634/stemcells.2008-0922. Stem Cells. 2009. PMID: 19259936 Free PMC article.
-
Characterization of human embryonic stem cell-derived hematopoietic progenitor phenotype.In Vitro Cell Dev Biol Anim. 2010 Oct;46(9):733-7. doi: 10.1007/s11626-010-9337-8. Epub 2010 Jul 10. In Vitro Cell Dev Biol Anim. 2010. PMID: 20623201
-
Human embryonic stem cell-derived CD34+ cells: efficient production in the coculture with OP9 stromal cells and analysis of lymphohematopoietic potential.Blood. 2005 Jan 15;105(2):617-26. doi: 10.1182/blood-2004-04-1649. Epub 2004 Sep 16. Blood. 2005. PMID: 15374881
-
Hematopoietic differentiation.2012 Jun 10. In: StemBook [Internet]. Cambridge (MA): Harvard Stem Cell Institute; 2008–. 2012 Jun 10. In: StemBook [Internet]. Cambridge (MA): Harvard Stem Cell Institute; 2008–. PMID: 23658971 Free Books & Documents. Review.
-
Update on the pathogenesis of Scleroderma: focus on circulating progenitor cells.F1000Res. 2016 Apr 22;5:F1000 Faculty Rev-723. doi: 10.12688/f1000research.7986.1. eCollection 2016. F1000Res. 2016. PMID: 27158466 Free PMC article. Review.
Cited by
-
Generation of T cells from Human and Nonhuman Primate Pluripotent Stem Cells.Bio Protoc. 2020 Jul 5;10(13):e3675. doi: 10.21769/BioProtoc.3675. eCollection 2020 Jul 5. Bio Protoc. 2020. PMID: 33659345 Free PMC article.
-
The Notch ligand DLL4 specifically marks human hematoendothelial progenitors and regulates their hematopoietic fate.Leukemia. 2015 Aug;29(8):1741-53. doi: 10.1038/leu.2015.74. Epub 2015 Mar 17. Leukemia. 2015. PMID: 25778099
-
Generation of mature hematopoietic cells from human pluripotent stem cells.Int J Hematol. 2012 Jun;95(6):617-23. doi: 10.1007/s12185-012-1094-x. Epub 2012 May 31. Int J Hematol. 2012. PMID: 22648826 Review.
-
Medial HOXA genes demarcate haematopoietic stem cell fate during human development.Nat Cell Biol. 2016 Jun;18(6):595-606. doi: 10.1038/ncb3354. Epub 2016 May 16. Nat Cell Biol. 2016. PMID: 27183470 Free PMC article.
-
GFI1 regulates chromatin state essential in human endothelial-to-haematopoietic transition.Cell Prolif. 2022 May;55(5):e13244. doi: 10.1111/cpr.13244. Epub 2022 May 3. Cell Prolif. 2022. PMID: 35504619 Free PMC article.
References
-
- Godin I, Cumano A. The hare and the tortoise: an embryonic haematopoietic race. Nat Rev Immunol. 2002;2: 593-604. - PubMed
-
- Tavian M, Zheng B, Oberlin E, et al. The vascular wall as a source of stem cells. Ann N Y Acad Sci. 2005;1044: 41-50. - PubMed
-
- Li W, Ferkowicz MJ, Johnson SA, Shelley WC, Yoder MC. Endothelial cells in the early murine yolk sac give rise to CD41-expressing hematopoietic cells. Stem Cells Dev. 2005;14: 44-54. - PubMed
-
- Jaffredo T, Bollerot K, Sugiyama D, Gautier R, Drevon C. Tracing the hemangioblast during embryogenesis: developmental relationships between endothelial and hematopoietic cells. Int J Dev Biol. 2005;49: 269-277. - PubMed
-
- Fraser ST, Ogawa M, Yu RT, Nishikawa S, Yoder MC. Definitive hematopoietic commitment within the embryonic vascular endothelial-cadherin(+) population. Exp Hematol. 2002;30: 1070-1078. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

