Flanking sequence composition differentially affects the binding and functional characteristics of glucocorticoid receptor homo- and heterodimers
- PMID: 16752918
- PMCID: PMC2517624
- DOI: 10.1021/bi060314k
Flanking sequence composition differentially affects the binding and functional characteristics of glucocorticoid receptor homo- and heterodimers
Abstract
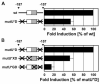
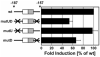
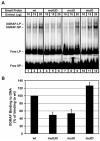
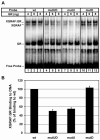
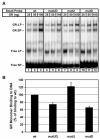
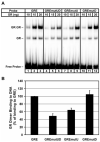
The core binding sites for a multitude of transcription factors have been identified and characterized, but these sequences cannot fully account for the nuances of cell-specific and gene-specific control of gene transcription. Many factors may contribute to the precise responsiveness of a gene to a particular transcriptional regulatory protein, including the nucleotides in the proximity of the core binding site for that protein. Here, we examine two flanking sequences bordering a site in the gamma-fibrinogen gene regulatory region that binds a heterodimer of the Xenopus glucocorticoid receptor accessory factor (XGRAF) and the glucocorticoid receptor (GR). Mutation of the upstream flank results in a decrease in the level of XGRAF binding but little change in hormone induction. However, alteration of the downstream flank adjacent to the GR binding site causes a decrease in levels of both GR monomer binding and hormone induction. Conversion of the XGRAF-GR binding site to a full glucocorticoid response element (GRE) alters the role of the flanking sequences. A full GRE in this position requires the wild-type upstream flank to bind GR homodimer and induce transcription to maximal levels. In contrast, mutation of the downstream flank is not detrimental to either the binding or the function of the GR dimer. Thus, flanking sequence composition and dimer partner combine to influence GR function, underscoring the complexities involved in the identification of authentic transcription factor response elements.
Figures


Similar articles
-
Heterodimerization between the glucocorticoid receptor and the unrelated DNA-binding protein, Xenopus glucocorticoid receptor accessory factor.Mol Endocrinol. 2001 Mar;15(3):458-66. doi: 10.1210/mend.15.3.0607. Mol Endocrinol. 2001. PMID: 11222746
-
The binding site for Xenopus glucocorticoid receptor accessory factor and a single adjacent half-GRE form an independent glucocorticoid response unit.Biochemistry. 2000 Oct 10;39(40):12234-42. doi: 10.1021/bi000981s. Biochemistry. 2000. PMID: 11015202
-
Analysis of the DNA-binding site for Xenopus glucocorticoid receptor accessory factor. Critical nucleotides for binding specificity in vitro and for amplification of steroid-induced fibrinogen gene transcription.J Biol Chem. 1998 Apr 17;273(16):9790-6. doi: 10.1074/jbc.273.16.9790. J Biol Chem. 1998. PMID: 9545317
-
How glucocorticoid receptors modulate the activity of other transcription factors: a scope beyond tethering.Mol Cell Endocrinol. 2013 Nov 5;380(1-2):41-54. doi: 10.1016/j.mce.2012.12.014. Epub 2012 Dec 23. Mol Cell Endocrinol. 2013. PMID: 23267834 Review.
-
New insights into the anti-inflammatory mechanisms of glucocorticoids: an emerging role for glucocorticoid-receptor-mediated transactivation.Endocrinology. 2013 Mar;154(3):993-1007. doi: 10.1210/en.2012-2045. Epub 2013 Feb 5. Endocrinology. 2013. PMID: 23384835 Review.
Cited by
-
Glucocorticoid therapy and ocular hypertension.Eur J Pharmacol. 2016 Sep 15;787:57-71. doi: 10.1016/j.ejphar.2016.06.018. Epub 2016 Jul 5. Eur J Pharmacol. 2016. PMID: 27388141 Free PMC article. Review.
-
Genomic regions flanking E-box binding sites influence DNA binding specificity of bHLH transcription factors through DNA shape.Cell Rep. 2013 Apr 25;3(4):1093-104. doi: 10.1016/j.celrep.2013.03.014. Epub 2013 Apr 4. Cell Rep. 2013. PMID: 23562153 Free PMC article.
-
CTCF binding site sequence differences are associated with unique regulatory and functional trends during embryonic stem cell differentiation.Nucleic Acids Res. 2014 Jan;42(2):774-89. doi: 10.1093/nar/gkt910. Epub 2013 Oct 10. Nucleic Acids Res. 2014. PMID: 24121688 Free PMC article.
-
Using protein-binding microarrays to study transcription factor specificity: homologs, isoforms and complexes.Brief Funct Genomics. 2015 Jan;14(1):17-29. doi: 10.1093/bfgp/elu046. Epub 2014 Nov 26. Brief Funct Genomics. 2015. PMID: 25431149 Free PMC article. Review.
-
Genome-wide analysis of AAAG and ACGT cis-elements in Arabidopsis thaliana reveals their involvement with genes downregulated under jasmonic acid response in an orientation independent manner.G3 (Bethesda). 2022 May 6;12(5):jkac057. doi: 10.1093/g3journal/jkac057. G3 (Bethesda). 2022. PMID: 35302624 Free PMC article.
References
-
- Lefstin JA, Yamamoto KR. Allosteric effects of DNA on transcriptional regulators. Nature. 1998;392:885–888. - PubMed
-
- Leung TH, Hoffmann A, Baltimore D. One nucleotide in a κB site can determine cofactor specificity for NF-κB dimers. Cell. 2004;118:453–464. - PubMed
-
- Natoli G. Little things that count in transcriptional regulation. Cell. 2004;118:406–408. - PubMed
-
- Mangelsdorf DJ, Evans RM. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

