Evaluation of methods for sequence analysis of highly repetitive DNA templates
- PMID: 16741241
- PMCID: PMC2291771
Evaluation of methods for sequence analysis of highly repetitive DNA templates
Abstract
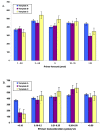
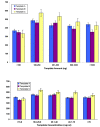
The DNA Sequencing Research Group (DSRG) of the ABRF conducted a study to assess the ability of DNA sequencing core facilities to successfully sequence a set of well-defined templates containing difficult repeats. The aim of this study was to determine whether repetitive templates could be sequenced accurately by using equipment and chemistries currently utilized in participating sequencing laboratories. The effects of primer and template concentrations, sequencing chemistries, additives, and instrument formats on the ability to successfully sequence repeat elements were examined. The first part of this study was an analysis of the results of 361 chromatograms from participants representing 40 different laboratories who attempted to sequence a panel of difficult-to-sequence templates using their best in-house protocols. The second part of this study was a smaller multi-laboratory evaluation of a single robust protocol with the same panel of templates. This study provides a measure of the potential success of different approaches to sequencing across homopolymer tracts and repetitive elements.
Figures

Similar articles
-
Identification of optimal protocols for sequencing difficult templates: results of the 2008 ABRF DNA Sequencing Research Group difficult template study 2008.J Biomol Tech. 2009 Apr;20(2):116-27. J Biomol Tech. 2009. PMID: 19503623 Free PMC article.
-
Accuracy of automated DNA sequencing: a multi-laboratory comparison of sequencing results.Biotechniques. 1995 Sep;19(3):448-53. Biotechniques. 1995. PMID: 7495559
-
DNA sequencing by synthesis with degenerate primers.J Genet Genomics. 2008 Sep;35(9):545-51. doi: 10.1016/S1673-8527(08)60074-0. J Genet Genomics. 2008. PMID: 18804073
-
Primer design and primer-directed sequencing.Methods Mol Biol. 2001;167:39-51. doi: 10.1385/1-59259-113-2:039. Methods Mol Biol. 2001. PMID: 11265320 Review. No abstract available.
-
The new paradigm of flow cell sequencing.Genome Res. 2008 Jun;18(6):839-46. doi: 10.1101/gr.073262.107. Genome Res. 2008. PMID: 18519653 Review.
Cited by
-
A Review of the Scientific Rigor, Reproducibility, and Transparency Studies Conducted by the ABRF Research Groups.J Biomol Tech. 2020 Apr;31(1):11-26. doi: 10.7171/jbt.20-3101-003. J Biomol Tech. 2020. PMID: 31969795 Free PMC article. Review.
-
Mitochondrial DNA Analysis.Methods Mol Biol. 2023;2685:331-349. doi: 10.1007/978-1-0716-3295-6_20. Methods Mol Biol. 2023. PMID: 37439991
References
-
- Lewin B. Genes VI. New York: Oxford University Press, Inc. 1997.
-
- Robbins CM, Hsu E, Gillevet PM. Sequencing homopolymer tracts and repetitive elements. BioTechniques 1996;20:862–868. - PubMed
-
- Langan JE, Rowbottom L, Liloglou T, Field JK, Risk JM. Sequencing of difficult templates containing poly(A/T) tracts: Closure of sequence gaps. BioTechniques 2002;33:276–280. - PubMed
-
- Stirling D. Technical notes for sequencing difficult templates. Methods Mol Biol 2003;226:401–402. - PubMed
-
- Wiebe GJ, Pershad R, Escobar H, Hawes JW, Hunter T, Jackson-Machelski E, et al. DNA Sequencing Research Group (DSRG) 2003—A general survey of core DNA sequencing facilities. J Biomol Tech 2003;14:230–236.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
