Regulation of mitochondrial fusion by the F-box protein Mdm30 involves proteasome-independent turnover of Fzo1
- PMID: 16735578
- PMCID: PMC2063881
- DOI: 10.1083/jcb.200512079
Regulation of mitochondrial fusion by the F-box protein Mdm30 involves proteasome-independent turnover of Fzo1
Abstract
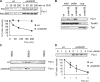
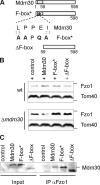
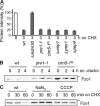
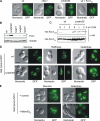
Mitochondrial morphology depends on balanced fusion and fission events. A central component of the mitochondrial fusion apparatus is the conserved GTPase Fzo1 in the outer membrane of mitochondria. Mdm30, an F-box protein required for mitochondrial fusion in vegetatively growing cells, affects the cellular Fzo1 concentration in an unknown manner. We demonstrate that mitochondrial fusion requires a tight control of Fzo1 levels, which is ensured by Fzo1 turnover. Mdm30 binds to Fzo1 and, dependent on its F-box, mediates proteolysis of Fzo1. Unexpectedly, degradation occurs along a novel proteolytic pathway not involving ubiquitylation, Skp1-Cdc53-F-box (SCF) E3 ubiquitin ligase complexes, or 26S proteasomes, indicating a novel function of an F-box protein. This contrasts to the ubiquitin- and proteasome-dependent turnover of Fzo1 in alpha-factor-arrested yeast cells. Our results therefore reveal not only a critical role of Fzo1 degradation for mitochondrial fusion in vegetatively growing cells but also the existence of two distinct proteolytic pathways for the turnover of mitochondrial outer membrane proteins.
Figures

Similar articles
-
Sequential requirements for the GTPase domain of the mitofusin Fzo1 and the ubiquitin ligase SCFMdm30 in mitochondrial outer membrane fusion.J Cell Sci. 2011 May 1;124(Pt 9):1403-10. doi: 10.1242/jcs.079293. J Cell Sci. 2011. PMID: 21502136 Free PMC article.
-
Mdm30 is an F-box protein required for maintenance of fusion-competent mitochondria in yeast.Mol Biol Cell. 2003 Jun;14(6):2303-13. doi: 10.1091/mbc.e02-12-0831. Epub 2003 Feb 6. Mol Biol Cell. 2003. PMID: 12808031 Free PMC article.
-
Ugo1 and Mdm30 act sequentially during Fzo1-mediated mitochondrial outer membrane fusion.J Cell Sci. 2011 Apr 1;124(Pt 7):1126-35. doi: 10.1242/jcs.073080. Epub 2011 Mar 8. J Cell Sci. 2011. PMID: 21385840
-
Outer mitochondrial membrane protein degradation by the proteasome.Novartis Found Symp. 2007;287:4-14; discussion 14-20. Novartis Found Symp. 2007. PMID: 18074628 Review.
-
Proteolytic regulation of metabolic enzymes by E3 ubiquitin ligase complexes: lessons from yeast.Crit Rev Biochem Mol Biol. 2015;50(6):489-502. doi: 10.3109/10409238.2015.1081869. Epub 2015 Sep 11. Crit Rev Biochem Mol Biol. 2015. PMID: 26362128 Review.
Cited by
-
A Tale of Two Proteolytic Machines: Matrix Metalloproteinases and the Ubiquitin-Proteasome System in Pulmonary Fibrosis.Int J Mol Sci. 2020 May 29;21(11):3878. doi: 10.3390/ijms21113878. Int J Mol Sci. 2020. PMID: 32485920 Free PMC article. Review.
-
Regulation of Mitochondrial Dynamics by Proteolytic Processing and Protein Turnover.Antioxidants (Basel). 2018 Jan 17;7(1):15. doi: 10.3390/antiox7010015. Antioxidants (Basel). 2018. PMID: 29342083 Free PMC article. Review.
-
Schizosaccharomyces pombe Fzo1 is subjected to the ubiquitin-proteasome-mediated degradation during the stationary phase.Int Microbiol. 2022 May;25(2):397-404. doi: 10.1007/s10123-022-00231-2. Epub 2022 Jan 25. Int Microbiol. 2022. PMID: 35075549
-
Mitochondrial Surveillance by Cdc48/p97: MAD vs. Membrane Fusion.Int J Mol Sci. 2020 Sep 18;21(18):6841. doi: 10.3390/ijms21186841. Int J Mol Sci. 2020. PMID: 32961852 Free PMC article. Review.
-
Tetratricopeptide repeat proteins Tom70 and Tom71 mediate yeast mitochondrial morphogenesis.EMBO Rep. 2008 Jan;9(1):63-9. doi: 10.1038/sj.embor.7401113. Epub 2007 Nov 16. EMBO Rep. 2008. PMID: 18007655 Free PMC article.
References
-
- Chen, H., and D.C. Chan. 2005. Emerging functions of mammalian mitochondrial fusion and fission. Hum. Mol. Genet. 14:R283–R289. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

