Positioning of proteins in membranes: a computational approach
- PMID: 16731967
- PMCID: PMC2242528
- DOI: 10.1110/ps.062126106
Positioning of proteins in membranes: a computational approach
Abstract
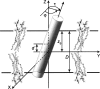
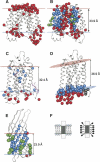
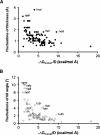
A new computational approach has been developed to determine the spatial arrangement of proteins in membranes by minimizing their transfer energies from water to the lipid bilayer. The membrane hydrocarbon core was approximated as a planar slab of adjustable thickness with decadiene-like interior and interfacial polarity profiles derived from published EPR studies. Applicability and accuracy of the method was verified for a set of 24 transmembrane proteins whose orientations in membranes have been studied by spin-labeling, chemical modification, fluorescence, ATR FTIR, NMR, cryo-microscopy, and neutron diffraction. Subsequently, the optimal rotational and translational positions were calculated for 109 transmembrane, five integral monotopic and 27 peripheral protein complexes with known 3D structures. This method can reliably distinguish transmembrane and integral monotopic proteins from water-soluble proteins based on their transfer energies and membrane penetration depths. The accuracies of calculated hydrophobic thicknesses and tilt angles were approximately 1 A and 2 degrees, respectively, judging from their deviations in different crystal forms of the same proteins. The hydrophobic thicknesses of transmembrane proteins ranged from 21.1 to 43.8 A depending on the type of biological membrane, while their tilt angles with respect to the bilayer normal varied from zero in symmetric complexes to 26 degrees in asymmetric structures. Calculated hydrophobic boundaries of proteins are located approximately 5 A lower than lipid phosphates and correspond to the zero membrane depth parameter of spin-labeled residues. Coordinates of all studied proteins with their membrane boundaries can be found in the Orientations of Proteins in Membranes (OPM) database:http://opm.phar.umich.edu/.
Figures


Similar articles
-
The role of hydrophobic interactions in positioning of peripheral proteins in membranes.BMC Struct Biol. 2007 Jun 29;7:44. doi: 10.1186/1472-6807-7-44. BMC Struct Biol. 2007. PMID: 17603894 Free PMC article.
-
OPM: orientations of proteins in membranes database.Bioinformatics. 2006 Mar 1;22(5):623-5. doi: 10.1093/bioinformatics/btk023. Epub 2006 Jan 5. Bioinformatics. 2006. PMID: 16397007
-
Anisotropic solvent model of the lipid bilayer. 2. Energetics of insertion of small molecules, peptides, and proteins in membranes.J Chem Inf Model. 2011 Apr 25;51(4):930-46. doi: 10.1021/ci200020k. Epub 2011 Mar 25. J Chem Inf Model. 2011. PMID: 21438606 Free PMC article.
-
Bilayer hydrophobic thickness and integral membrane protein function.Curr Protein Pept Sci. 2011 Dec;12(8):760-6. doi: 10.2174/138920311798841681. Curr Protein Pept Sci. 2011. PMID: 22044142 Review.
-
Life at the border: adaptation of proteins to anisotropic membrane environment.Protein Sci. 2014 Sep;23(9):1165-96. doi: 10.1002/pro.2508. Epub 2014 Jul 2. Protein Sci. 2014. PMID: 24947665 Free PMC article. Review.
Cited by
-
Structural and mechanistic analysis of the membrane-embedded glycosyltransferase WaaA required for lipopolysaccharide synthesis.Proc Natl Acad Sci U S A. 2012 Apr 17;109(16):6253-8. doi: 10.1073/pnas.1119894109. Epub 2012 Apr 2. Proc Natl Acad Sci U S A. 2012. PMID: 22474366 Free PMC article.
-
Ceramide-1-phosphate transfer protein promotes sphingolipid reorientation needed for binding during membrane interaction.J Lipid Res. 2022 Jan;63(1):100151. doi: 10.1016/j.jlr.2021.100151. Epub 2021 Nov 20. J Lipid Res. 2022. PMID: 34808193 Free PMC article.
-
The mechanism of sodium and substrate release from the binding pocket of vSGLT.Nature. 2010 Dec 16;468(7326):988-91. doi: 10.1038/nature09580. Epub 2010 Dec 5. Nature. 2010. PMID: 21131949 Free PMC article.
-
Orientation of the Escherichia coli outer membrane protein OmpX in phospholipid bilayer membranes determined by solid-State NMR.Biochemistry. 2008 Jun 24;47(25):6531-8. doi: 10.1021/bi800362b. Biochemistry. 2008. PMID: 18512961 Free PMC article.
-
Identification of interaction partners of outer inflammatory protein A: Computational and experimental insights into how Helicobacter pylori infects host cells.PLoS One. 2024 Oct 29;19(10):e0300557. doi: 10.1371/journal.pone.0300557. eCollection 2024. PLoS One. 2024. PMID: 39471168 Free PMC article.
References
-
- Abraham M.H., Chadha H.S., Whiting G.S., Mitchell R.C. 1994. Hydrogen bonding. 32. An analysis of water-octanol and water-alkane partitioning and the ΔlogP parameter of Seiler J. Pharm. Sci. 83 1085–1100. - PubMed
-
- Altenbach C., Marti T., Khorana H.G., Hubbell W.L. 1990. Transmembrane protein structure: spin labeling of bacteriorhodopsin mutants Science 248 1088–1092. - PubMed
-
- Andersen O.S., Koeppe R.E., Roux B. 2005. Gramicidin channels IEEE Trans Nanobioscience 4 10–20. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

