Autoimmune arthritis associated with mutated interleukin (IL)-6 receptor gp130 is driven by STAT3/IL-7-dependent homeostatic proliferation of CD4+ T cells
- PMID: 16717113
- PMCID: PMC2118324
- DOI: 10.1084/jem.20052187
Autoimmune arthritis associated with mutated interleukin (IL)-6 receptor gp130 is driven by STAT3/IL-7-dependent homeostatic proliferation of CD4+ T cells
Abstract
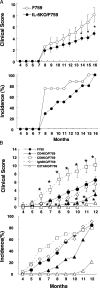
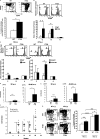
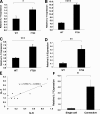
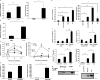
Mice homozygous for the F759 mutation in the gp130 interleukin (IL)-6 receptor subunit have enhanced gp130-mediated signal transducer and activator of transcription (STAT)3 activation and spontaneously developed a lymphocyte-mediated rheumatoid arthritis-like joint disease. Here, we show that the development of the disease is dependent on both major histocompatibility complex (MHC) II-restricted CD4+ T cells and IL-6 family cytokines. In spite of the necessity for CD4+ T cells, the gp130 mutation was only required in nonhemtopoietic cells for the disease. The gp130 mutation resulted in enhanced production of IL-7. Conditional knockout of STAT3 in nonlymphoid cells showed that the enhancement of IL-7 production was dependent on STAT3 activation by IL-6 family cytokines. Homeostatic proliferation of CD4+ T cells was enhanced in gp130 mutant mice and acceleration of homeostatic proliferation enhanced the disease, whereas the inhibition of homeostatic proliferation suppressed the disease. Anti-IL-7 antibody treatment inhibited not only the enhanced homeostatic proliferation, but also the disease in gp130 mutant mice. Thus, our results show that autoimmune disease in gp130 mutant mice is caused by increased homeostatic proliferation of CD4+ T cells, which is due to elevated production of IL-7 by nonhematopoietic cells as a result of IL-6 family cytokine-gp130-STAT3 signaling.
Figures


Similar articles
-
IL-6-gp130-STAT3 in T cells directs the development of IL-17+ Th with a minimum effect on that of Treg in the steady state.Int Immunol. 2007 Jun;19(6):695-702. doi: 10.1093/intimm/dxm045. Epub 2007 May 9. Int Immunol. 2007. PMID: 17493959
-
A positive feedback loop of IL-21 signaling provoked by homeostatic CD4+CD25- T cell expansion is essential for the development of arthritis in autoimmune K/BxN mice.J Immunol. 2009 Apr 15;182(8):4649-56. doi: 10.4049/jimmunol.0804350. J Immunol. 2009. PMID: 19342640
-
IL-6 positively regulates Foxp3+CD8+ T cells in vivo.Int Immunol. 2010 Feb;22(2):129-39. doi: 10.1093/intimm/dxp119. Epub 2009 Dec 30. Int Immunol. 2010. PMID: 20042455
-
IL-6 signal transduction and its physiological roles: the signal orchestration model.Rev Physiol Biochem Pharmacol. 2003;149:1-38. doi: 10.1007/s10254-003-0012-2. Epub 2003 Apr 5. Rev Physiol Biochem Pharmacol. 2003. PMID: 12687404 Review.
-
[Abnormal gp130 signaling and autoimmune disease].Tanpakushitsu Kakusan Koso. 2002 Dec;47(16 Suppl):2336-42. Tanpakushitsu Kakusan Koso. 2002. PMID: 12518458 Review. Japanese. No abstract available.
Cited by
-
Live from the liver: hepatocyte IL-7.Immunity. 2009 Mar 20;30(3):320-1. doi: 10.1016/j.immuni.2009.03.001. Immunity. 2009. PMID: 19303385 Free PMC article.
-
IL-6 in inflammation, autoimmunity and cancer.Int Immunol. 2021 Mar 1;33(3):127-148. doi: 10.1093/intimm/dxaa078. Int Immunol. 2021. PMID: 33337480 Free PMC article. Review.
-
The role of all-trans retinoic acid in the biology of Foxp3+ regulatory T cells.Cell Mol Immunol. 2015 Sep;12(5):553-7. doi: 10.1038/cmi.2014.133. Epub 2015 Feb 2. Cell Mol Immunol. 2015. PMID: 25640656 Free PMC article. Review.
-
Human CD39hi regulatory T cells present stronger stability and function under inflammatory conditions.Cell Mol Immunol. 2017 Jun;14(6):521-528. doi: 10.1038/cmi.2016.30. Epub 2016 Jul 4. Cell Mol Immunol. 2017. PMID: 27374793 Free PMC article.
-
Characterization of a novel and spontaneous mouse model of inflammatory arthritis.Arthritis Res Ther. 2011 Jul 12;13(4):R114. doi: 10.1186/ar3399. Arthritis Res Ther. 2011. PMID: 21749708 Free PMC article.
References
-
- Firestein, G.S. 2003. Evolving concepts of rheumatoid arthritis. Nature. 423:356–361. - PubMed
-
- Marrack, P., J. Kappler, and B. Kotzin. 2001. Autoimmune disease: why and where it occurs. Nat. Med. 7:899–905. - PubMed
-
- Hirano, T. 2002. Cytokines in autoimmune disease and chronic inflammatory proliferative disease. Cytokine Growth Factor Rev. 13:297–298. - PubMed
-
- Kontoyiannis, D., M. Pasparakis, T. Pizarro, F. Cominelli, and G. Kollias. 1999. Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: implications for joint and gut-associated immunopathologies. Immunity. 10:387–398. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

