Topoisomerase II, not topoisomerase I, is the proficient relaxase of nucleosomal DNA
- PMID: 16710299
- PMCID: PMC1478187
- DOI: 10.1038/sj.emboj.7601142
Topoisomerase II, not topoisomerase I, is the proficient relaxase of nucleosomal DNA
Abstract
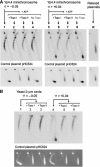
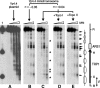
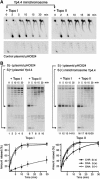
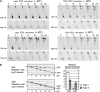
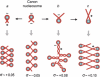
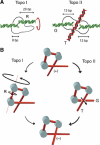
Eukaryotic topoisomerases I and II efficiently remove helical tension in naked DNA molecules. However, this activity has not been examined in nucleosomal DNA, their natural substrate. Here, we obtained yeast minichromosomes holding DNA under (+) helical tension, and incubated them with topoisomerases. We show that DNA supercoiling density can rise above +0.04 without displacement of the histones and that the typical nucleosome topology is restored upon DNA relaxation. However, in contrast to what is observed in naked DNA, topoisomerase II relaxes nucleosomal DNA much faster than topoisomerase I. The same effect occurs in cell extracts containing physiological dosages of topoisomeraseI and II. Apparently, the DNA strand-rotation mechanism of topoisomerase I does not efficiently relax chromatin, which imposes barriers for DNA twist diffusion. Conversely, the DNA cross-inversion mechanism of topoisomerase II is facilitated in chromatin, which favor the juxtaposition of DNA segments. We conclude that topoisomerase II is the main modulator of DNA topology in chromatin fibers. The nonessential topoisomerase I then assists DNA relaxation where chromatin structure impairs DNA juxtaposition but allows twist diffusion.
Figures

Similar articles
-
[DNA supercoiling and topoisomerases in Escherichia coli].Rev Latinoam Microbiol. 1995 Jul-Sep;37(3):291-304. Rev Latinoam Microbiol. 1995. PMID: 8850348 Review. Spanish.
-
Varying levels of positive and negative supercoiling differently affect the efficiency with which topoisomerase II catenates and decatenates DNA.J Mol Biol. 2001 Jan 19;305(3):441-50. doi: 10.1006/jmbi.2000.4307. J Mol Biol. 2001. PMID: 11152602
-
Transcriptional supercoiling boosts topoisomerase II-mediated knotting of intracellular DNA.Nucleic Acids Res. 2019 Jul 26;47(13):6946-6955. doi: 10.1093/nar/gkz491. Nucleic Acids Res. 2019. PMID: 31165864 Free PMC article.
-
Failure to relax negative supercoiling of DNA is a primary cause of mitotic hyper-recombination in topoisomerase-deficient yeast cells.J Biol Chem. 2002 Oct 4;277(40):37207-11. doi: 10.1074/jbc.M206663200. Epub 2002 Jul 31. J Biol Chem. 2002. PMID: 12151411
-
DNA supercoiling and relaxation by ATP-dependent DNA topoisomerases.Philos Trans R Soc Lond B Biol Sci. 1992 Apr 29;336(1276):83-91. doi: 10.1098/rstb.1992.0047. Philos Trans R Soc Lond B Biol Sci. 1992. PMID: 1351300 Review.
Cited by
-
A method for genome-wide analysis of DNA helical tension by means of psoralen-DNA photobinding.Nucleic Acids Res. 2010 Oct;38(19):e182. doi: 10.1093/nar/gkq687. Epub 2010 Aug 4. Nucleic Acids Res. 2010. PMID: 20685815 Free PMC article.
-
Human topoisomerases and their roles in genome stability and organization.Nat Rev Mol Cell Biol. 2022 Jun;23(6):407-427. doi: 10.1038/s41580-022-00452-3. Epub 2022 Feb 28. Nat Rev Mol Cell Biol. 2022. PMID: 35228717 Free PMC article. Review.
-
MGOUN1 encodes an Arabidopsis type IB DNA topoisomerase required in stem cell regulation and to maintain developmentally regulated gene silencing.Plant Cell. 2010 Mar;22(3):716-28. doi: 10.1105/tpc.109.068296. Epub 2010 Mar 12. Plant Cell. 2010. PMID: 20228247 Free PMC article.
-
Topoisomerase IIbeta activates a subset of neuronal genes that are repressed in AT-rich genomic environment.PLoS One. 2008;3(12):e4103. doi: 10.1371/journal.pone.0004103. Epub 2008 Dec 31. PLoS One. 2008. PMID: 19116664 Free PMC article.
-
Enzymatic Processing of DNA-Protein Crosslinks.Genes (Basel). 2024 Jan 10;15(1):85. doi: 10.3390/genes15010085. Genes (Basel). 2024. PMID: 38254974 Free PMC article. Review.
References
-
- Adachi Y, Luke M, Laemmli UK (1991) Chromosome assembly in vitro: topoisomerase II is required for condensation. Cell 64: 137–148 - PubMed
-
- Bednar J, Furrer P, Stasiak A, Dubochet J, Egelman EH, Bates AD (1994) The twist, writhe and overall shape of supercoiled DNA change during counterion-induced transition from a loosely to a tightly interwound superhelix. J Mol Biol 235: 825–847 - PubMed
-
- Bjornsti MA, Fertala J (1999) Overexpression and purification of DNA topoisomerase I from yeast. Methods Mol Biol 94: 179–186 - PubMed
-
- Boles TC, White JH, Cozzarelli NR (1990) Structure of plectonemically supercoiled DNA. J Mol Biol 213: 931–951 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

