The AAA ATPase p97 links peptide N-glycanase to the endoplasmic reticulum-associated E3 ligase autocrine motility factor receptor
- PMID: 16709668
- PMCID: PMC1482497
- DOI: 10.1073/pnas.0602747103
The AAA ATPase p97 links peptide N-glycanase to the endoplasmic reticulum-associated E3 ligase autocrine motility factor receptor
Abstract
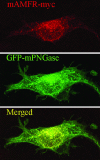
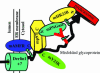
Mouse peptide N-glycanase (mPNGase) cleaves the N-glycan chain from misfolded glycoproteins and glycopeptides. Previously, several proteins were found to directly interact with mPNGase; among them, both mHR23B and mS4 were found to link mPNGase to the proteasome. In this study, we found that the cytoplasmic protein mp97 participates in the formation of a ternary complex containing mouse autocrine motility factor receptor (mAMFR), mp97, and mPNGase. This assemblage recruits the cytosolic mPNGase close to the endoplasmic reticulum (ER) membrane, where the retrotranslocation of misfolded glycoproteins is thought to occur. In addition to the ER membrane-associated E3 ligase mAMFR, a cytosolic protein mY33K, containing both UBA and UBX domains, was found to also directly interact with mp97. Thus, a complex containing five proteins, mAMFR, mY33K, mp97, mPNGase, and mHR23B, is formed in close proximity to the ER membrane and serves to couple the activities of retrotranslocation, ubiquitination, and deglycosylation and, thereby, route misfolded glycoproteins to the proteasome.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures




Similar articles
-
Multiple modes of interaction of the deglycosylation enzyme, mouse peptide N-glycanase, with the proteasome.Proc Natl Acad Sci U S A. 2005 Nov 1;102(44):15809-14. doi: 10.1073/pnas.0507155102. Epub 2005 Oct 25. Proc Natl Acad Sci U S A. 2005. PMID: 16249333 Free PMC article.
-
Regulation of ER-associated degradation via p97/VCP-interacting motif.Biochem Soc Trans. 2008 Oct;36(Pt 5):818-22. doi: 10.1042/BST0360818. Biochem Soc Trans. 2008. PMID: 18793143
-
Identification of SVIP as an endogenous inhibitor of endoplasmic reticulum-associated degradation.J Biol Chem. 2007 Nov 23;282(47):33908-14. doi: 10.1074/jbc.M704446200. Epub 2007 Sep 14. J Biol Chem. 2007. PMID: 17872946
-
The cytoplasmic peptide:N-glycanase (NGLY1) - Structure, expression and cellular functions.Gene. 2016 Feb 10;577(1):1-7. doi: 10.1016/j.gene.2015.11.021. Epub 2015 Nov 30. Gene. 2016. PMID: 26611529 Free PMC article. Review.
-
Physiological and molecular functions of the cytosolic peptide:N-glycanase.Semin Cell Dev Biol. 2015 May;41:110-20. doi: 10.1016/j.semcdb.2014.11.009. Epub 2014 Dec 2. Semin Cell Dev Biol. 2015. PMID: 25475175 Review.
Cited by
-
Dislocation of HMG-CoA reductase and Insig-1, two polytopic endoplasmic reticulum proteins, en route to proteasomal degradation.Mol Biol Cell. 2009 Jul;20(14):3330-41. doi: 10.1091/mbc.e08-09-0953. Epub 2009 May 20. Mol Biol Cell. 2009. PMID: 19458199 Free PMC article.
-
Protein Quality Control in the Endoplasmic Reticulum and Cancer.Int J Mol Sci. 2018 Oct 3;19(10):3020. doi: 10.3390/ijms19103020. Int J Mol Sci. 2018. PMID: 30282948 Free PMC article. Review.
-
The mammalian endoplasmic reticulum-associated degradation system.Cold Spring Harb Perspect Biol. 2013 Sep 1;5(9):a013185. doi: 10.1101/cshperspect.a013185. Cold Spring Harb Perspect Biol. 2013. PMID: 23232094 Free PMC article. Review.
-
A Mighty "Protein Extractor" of the Cell: Structure and Function of the p97/CDC48 ATPase.Front Mol Biosci. 2017 Jun 13;4:39. doi: 10.3389/fmolb.2017.00039. eCollection 2017. Front Mol Biosci. 2017. PMID: 28660197 Free PMC article. Review.
-
Defining human ERAD networks through an integrative mapping strategy.Nat Cell Biol. 2011 Nov 27;14(1):93-105. doi: 10.1038/ncb2383. Nat Cell Biol. 2011. PMID: 22119785 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

