The effects of multiple features of alternatively spliced exons on the K(A)/K(S) ratio test
- PMID: 16709259
- PMCID: PMC1526763
- DOI: 10.1186/1471-2105-7-259
The effects of multiple features of alternatively spliced exons on the K(A)/K(S) ratio test
Abstract
Background: The evolution of alternatively spliced exons (ASEs) is of primary interest because these exons are suggested to be a major source of functional diversity of proteins. Many exon features have been suggested to affect the evolution of ASEs. However, previous studies have relied on the KA/KS ratio test without taking into consideration information sufficiency (i.e., exon length > 75 bp, cross-species divergence > 5%) of the studied exons, leading to potentially biased interpretations. Furthermore, which exon feature dominates the results of the KA/KS ratio test and whether multiple exon features have additive effects have remained unexplored.
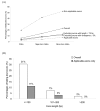
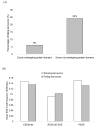
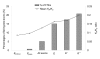
Results: In this study, we collect two different datasets for analysis - the ASE dataset (which includes lineage-specific ASEs and conserved ASEs) and the ACE dataset (which includes only conserved ASEs). We first show that information sufficiency can significantly affect the interpretation of relationship between exons features and the KA/KS ratio test results. After discarding exons with insufficient information, we use a Boolean method to analyze the relationship between test results and four exon features (namely length, protein domain overlapping, inclusion level, and exonic splicing enhancer (ESE) frequency) for the ASE dataset. We demonstrate that length and protein domain overlapping are dominant factors, and they have similar impacts on test results of ASEs. In addition, despite the weak impacts of inclusion level and ESE motif frequency when considered individually, combination of these two factors still have minor additive effects on test results. However, the ACE dataset shows a slightly different result in that inclusion level has a marginally significant effect on test results. Lineage-specific ASEs may have contributed to the difference. Overall, in both ASEs and ACEs, protein domain overlapping is the most dominant exon feature while ESE frequency is the weakest one in affecting test results.
Conclusion: The proposed method can easily find additive effects of individual or multiple factors on the KA/KS ratio test results of exons. Therefore, the system can analyze complex conditions in evolution where multiple features are involved. More factors can also be added into the system to extend the scope of evolutionary analysis of exons. In addition, our method may be useful when orthologous exons can not be found for the KA/KS ratio test.
Figures
Similar articles
-
Assessing the application of Ka/Ks ratio test to alternatively spliced exons.Bioinformatics. 2005 Oct 1;21(19):3701-3. doi: 10.1093/bioinformatics/bti613. Epub 2005 Aug 9. Bioinformatics. 2005. PMID: 16091412
-
Alternatively and constitutively spliced exons are subject to different evolutionary forces.Mol Biol Evol. 2006 Mar;23(3):675-82. doi: 10.1093/molbev/msj081. Epub 2005 Dec 20. Mol Biol Evol. 2006. PMID: 16368777
-
Opposite evolutionary effects between different alternative splicing patterns.Mol Biol Evol. 2007 Jul;24(7):1443-6. doi: 10.1093/molbev/msm072. Epub 2007 Apr 13. Mol Biol Evol. 2007. PMID: 17434901
-
The "alternative" choice of constitutive exons throughout evolution.PLoS Genet. 2007 Nov;3(11):e203. doi: 10.1371/journal.pgen.0030203. PLoS Genet. 2007. PMID: 18020709 Free PMC article.
-
How prevalent is functional alternative splicing in the human genome?Trends Genet. 2004 Feb;20(2):68-71. doi: 10.1016/j.tig.2003.12.004. Trends Genet. 2004. PMID: 14746986 Review.
Cited by
-
Different alternative splicing patterns are subject to opposite selection pressure for protein reading frame preservation.BMC Evol Biol. 2007 Sep 28;7:179. doi: 10.1186/1471-2148-7-179. BMC Evol Biol. 2007. PMID: 17900372 Free PMC article.
-
Position-dependent correlations between DNA methylation and the evolutionary rates of mammalian coding exons.Proc Natl Acad Sci U S A. 2012 Sep 25;109(39):15841-6. doi: 10.1073/pnas.1208214109. Epub 2012 Sep 10. Proc Natl Acad Sci U S A. 2012. PMID: 23019368 Free PMC article.
-
The relationships among microRNA regulation, intrinsically disordered regions, and other indicators of protein evolutionary rate.Mol Biol Evol. 2011 Sep;28(9):2513-20. doi: 10.1093/molbev/msr068. Epub 2011 Mar 11. Mol Biol Evol. 2011. PMID: 21398349 Free PMC article.
-
The evolution of the coding exome of the Arabidopsis species--the influences of DNA methylation, relative exon position, and exon length.BMC Evol Biol. 2014 Jun 25;14:145. doi: 10.1186/1471-2148-14-145. BMC Evol Biol. 2014. PMID: 24965500 Free PMC article.
-
Identification of a new susceptibility variant for multiple sclerosis in OAS1 by population genetics analysis.Hum Genet. 2012 Jan;131(1):87-97. doi: 10.1007/s00439-011-1053-2. Epub 2011 Jul 7. Hum Genet. 2012. PMID: 21735172 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

