The Fat1 cadherin integrates vascular smooth muscle cell growth and migration signals
- PMID: 16682528
- PMCID: PMC2063842
- DOI: 10.1083/jcb.200508121
The Fat1 cadherin integrates vascular smooth muscle cell growth and migration signals
Abstract
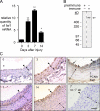
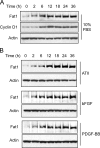
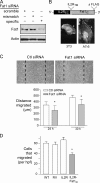
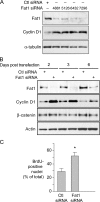
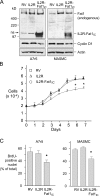
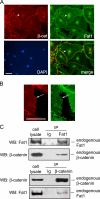
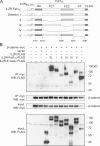
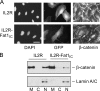
The significance of cadherin superfamily proteins in vascular smooth muscle cell (VSMC) biology is undefined. Here we describe recent studies of the Fat1 protocadherin. Fat1 expression in VSMCs increases significantly after arterial injury or growth factor stimulation. Fat1 knockdown decreases VSMC migration in vitro, but surprisingly, enhances cyclin D1 expression and proliferation. Despite limited similarity to classical cadherins, the Fat1 intracellular domain (Fat1(IC)) interacts with beta-catenin, inhibiting both its nuclear localization and transcriptional activity. Fat1 undergoes cleavage and Fat1(IC) species localize to the nucleus; however, inhibition of the cyclin D1 promoter by truncated Fat1(IC) proteins corresponds to their presence outside the nucleus, which argues against repression of beta-catenin-dependent transcription by nuclear Fat1(IC). These findings extend recent observations about Fat1 and migration in other cell types, and demonstrate for the first time its anti-proliferative activity and interaction with beta-catenin. Because it is induced after arterial injury, Fat1 may control VSMC functions central to vascular remodeling by facilitating migration and limiting proliferation.
Figures
Similar articles
-
R-cadherin:beta-catenin complex and its association with vascular smooth muscle cell proliferation.Arterioscler Thromb Vasc Biol. 2004 Jul;24(7):1204-10. doi: 10.1161/01.ATV.0000130464.24599.e0. Epub 2004 Apr 29. Arterioscler Thromb Vasc Biol. 2004. PMID: 15117735
-
Atrophin proteins interact with the Fat1 cadherin and regulate migration and orientation in vascular smooth muscle cells.J Biol Chem. 2009 Mar 13;284(11):6955-65. doi: 10.1074/jbc.M809333200. Epub 2009 Jan 7. J Biol Chem. 2009. PMID: 19131340 Free PMC article.
-
The FAT1 Cadherin Drives Vascular Smooth Muscle Cell Migration.Cells. 2023 Jun 14;12(12):1621. doi: 10.3390/cells12121621. Cells. 2023. PMID: 37371091 Free PMC article. Review.
-
MMP-9 and -12 cause N-cadherin shedding and thereby beta-catenin signalling and vascular smooth muscle cell proliferation.Cardiovasc Res. 2009 Jan 1;81(1):178-86. doi: 10.1093/cvr/cvn278. Epub 2008 Oct 13. Cardiovasc Res. 2009. PMID: 18852254
-
Regulation of VSMC behavior by the cadherin-catenin complex.Front Biosci (Landmark Ed). 2011 Jan 1;16(2):644-57. doi: 10.2741/3711. Front Biosci (Landmark Ed). 2011. PMID: 21196194 Review.
Cited by
-
Discoidin domain receptor 1 deficiency in vascular smooth muscle cells leads to mislocalisation of N-cadherin contacts.Biol Open. 2019 Aug 9;8(8):bio041913. doi: 10.1242/bio.041913. Biol Open. 2019. PMID: 31362952 Free PMC article.
-
Whole-exome sequencing in splenic marginal zone lymphoma reveals mutations in genes involved in marginal zone differentiation.Leukemia. 2014 Jun;28(6):1334-40. doi: 10.1038/leu.2013.365. Epub 2013 Dec 3. Leukemia. 2014. PMID: 24296945
-
A Subset of Autism-Associated Genes Regulate the Structural Stability of Neurons.Front Cell Neurosci. 2016 Nov 17;10:263. doi: 10.3389/fncel.2016.00263. eCollection 2016. Front Cell Neurosci. 2016. PMID: 27909399 Free PMC article. Review.
-
The role of T-cells in head and neck squamous cell carcinoma: From immunity to immunotherapy.Front Oncol. 2022 Oct 20;12:1021609. doi: 10.3389/fonc.2022.1021609. eCollection 2022. Front Oncol. 2022. PMID: 36338731 Free PMC article. Review.
-
β-Catenin C-terminal signals suppress p53 and are essential for artery formation.Nat Commun. 2016 Aug 8;7:12389. doi: 10.1038/ncomms12389. Nat Commun. 2016. PMID: 27499244 Free PMC article.
References
-
- Angst, B.D., C. Marcozzi, and A.I. Magee. 2001. The cadherin superfamily: diversity in form and function. J. Cell Sci. 114:629–641. - PubMed
-
- Bhanot, P., M. Fish, J.A. Jemison, R. Nusse, J. Nathans, and K.M. Cadigan. 1999. Frizzled and Dfrizzled-2 function as redundant receptors for Wingless during Drosophila embryonic development. Development. 126:4175–4186. - PubMed
-
- Bustin, S.A. 2000. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 25:169–193. - PubMed
-
- Castillejo-Lopez, C., W.M. Arias, and S. Baumgartner. 2004. The fat-like gene of Drosophila is the true orthologue of vertebrate fat cadherins and is involved in the formation of tubular organs. J. Biol. Chem. 279:24034–24043. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

