A structural model for monastrol inhibition of dimeric kinesin Eg5
- PMID: 16642039
- PMCID: PMC1462975
- DOI: 10.1038/sj.emboj.7601108
A structural model for monastrol inhibition of dimeric kinesin Eg5
Abstract
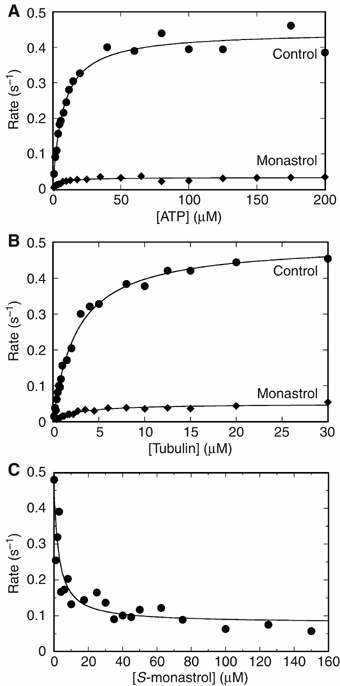
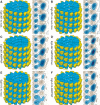
Eg5 or KSP is a homotetrameric Kinesin-5 involved in centrosome separation and assembly of the bipolar mitotic spindle. Analytical gel filtration of purified protein and cryo-electron microscopy (cryo-EM) of unidirectional shadowed microtubule-Eg5 complexes have been used to identify the stable dimer Eg5-513. The motility assays show that Eg5-513 promotes robust plus-end-directed microtubule gliding at a rate similar to that of homotetrameric Eg5 in vitro. Eg5-513 exhibits slow ATP turnover, high affinity for ATP, and a weakened affinity for microtubules when compared to monomeric Eg5. We show here that the Eg5-513 dimer binds microtubules with both heads to two adjacent tubulin heterodimers along the same microtubule protofilament. Under all nucleotide conditions tested, there were no visible structural changes in the monomeric Eg5-microtubule complexes with monastrol treatment. In contrast, there was a substantial monastrol effect on dimeric Eg5-513, which reduced microtubule lattice decoration. Comparisons between the X-ray structures of Eg5-ADP and Eg5-ADP-monastrol with rat kinesin-ADP after docking them into cryo-EM 3-D scaffolds revealed structural evidence for the weaker microtubule-Eg5 interaction in the presence of monastrol.
Figures


Similar articles
-
Monastrol inhibition of the mitotic kinesin Eg5.J Biol Chem. 2005 Apr 1;280(13):12658-67. doi: 10.1074/jbc.M413140200. Epub 2005 Jan 23. J Biol Chem. 2005. PMID: 15665380 Free PMC article.
-
Disparity in allosteric interactions of monastrol with Eg5 in the presence of ADP and ATP: a difference FT-IR investigation.Biochemistry. 2004 Aug 10;43(31):9939-49. doi: 10.1021/bi048982y. Biochemistry. 2004. PMID: 15287721
-
Interaction of the mitotic inhibitor monastrol with human kinesin Eg5.Biochemistry. 2003 Jan 21;42(2):338-49. doi: 10.1021/bi026716j. Biochemistry. 2003. PMID: 12525161
-
Eg5 targeting agents: From new anti-mitotic based inhibitor discovery to cancer therapy and resistance.Biochem Pharmacol. 2021 Feb;184:114364. doi: 10.1016/j.bcp.2020.114364. Epub 2020 Dec 11. Biochem Pharmacol. 2021. PMID: 33310050 Review.
-
Kinesin, 30 years later: Recent insights from structural studies.Protein Sci. 2015 Jul;24(7):1047-56. doi: 10.1002/pro.2697. Epub 2015 Jun 11. Protein Sci. 2015. PMID: 25975756 Free PMC article. Review.
Cited by
-
Force and premature binding of ADP can regulate the processivity of individual Eg5 dimers.Biophys J. 2009 Sep 16;97(6):1671-7. doi: 10.1016/j.bpj.2009.07.013. Biophys J. 2009. PMID: 19751672 Free PMC article.
-
Getting in sync with dimeric Eg5. Initiation and regulation of the processive run.J Biol Chem. 2008 Jan 25;283(4):2078-87. doi: 10.1074/jbc.M708354200. Epub 2007 Nov 25. J Biol Chem. 2008. PMID: 18037705 Free PMC article.
-
The structural basis of force generation by the mitotic motor kinesin-5.J Biol Chem. 2012 Dec 28;287(53):44654-66. doi: 10.1074/jbc.M112.404228. Epub 2012 Nov 7. J Biol Chem. 2012. PMID: 23135273 Free PMC article.
-
Pathway of ATP hydrolysis by monomeric kinesin Eg5.Biochemistry. 2006 Oct 10;45(40):12334-44. doi: 10.1021/bi0608562. Biochemistry. 2006. PMID: 17014086 Free PMC article.
-
Small molecule allosteric uncoupling of microtubule depolymerase activity from motility in human Kinesin-5 during mitotic spindle assembly.Sci Rep. 2019 Dec 27;9(1):19900. doi: 10.1038/s41598-019-56173-9. Sci Rep. 2019. PMID: 31882607 Free PMC article.
References
-
- Beuron F, Hoenger A (2001) Structural analysis of the microtubule–kinesin complex by cryo-electron microscopy. Methods Mol Biol 164: 235–254 - PubMed
-
- Blangy A, Lane HA, d'Herin P, Harper M, Kress M, Nigg EA (1995) Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell 83: 1159–1169 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

