Genome of crocodilepox virus
- PMID: 16641289
- PMCID: PMC1472061
- DOI: 10.1128/JVI.80.10.4978-4991.2006
Genome of crocodilepox virus
Abstract
Here, we present the genome sequence, with analysis, of a poxvirus infecting Nile crocodiles (Crocodylus niloticus) (crocodilepox virus; CRV). The genome is 190,054 bp (62% G+C) and predicted to contain 173 genes encoding proteins of 53 to 1,941 amino acids. The central genomic region contains genes conserved and generally colinear with those of other chordopoxviruses (ChPVs). CRV is distinct, as the terminal 33-kbp (left) and 13-kbp (right) genomic regions are largely CRV specific, containing 48 unique genes which lack similarity to other poxvirus genes. Notably, CRV also contains 14 unique genes which disrupt ChPV gene colinearity within the central genomic region, including 7 genes encoding GyrB-like ATPase domains similar to those in cellular type IIA DNA topoisomerases, suggestive of novel ATP-dependent functions. The presence of 10 CRV proteins with similarity to components of cellular multisubunit E3 ubiquitin-protein ligase complexes, including 9 proteins containing F-box motifs and F-box-associated regions and a homologue of cellular anaphase-promoting complex subunit 11 (Apc11), suggests that modification of host ubiquitination pathways may be significant for CRV-host cell interaction. CRV encodes a novel complement of proteins potentially involved in DNA replication, including a NAD(+)-dependent DNA ligase and a protein with similarity to both vaccinia virus F16L and prokaryotic serine site-specific resolvase-invertases. CRV lacks genes encoding proteins for nucleotide metabolism. CRV shares notable genomic similarities with molluscum contagiosum virus, including genes found only in these two viruses. Phylogenetic analysis indicates that CRV is quite distinct from other ChPVs, representing a new genus within the subfamily Chordopoxvirinae, and it lacks recognizable homologues of most ChPV genes involved in virulence and host range, including those involving interferon response, intracellular signaling, and host immune response modulation. These data reveal the unique nature of CRV and suggest mechanisms of virus-reptile host interaction.
Figures

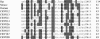


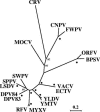
Similar articles
-
Molecular characterization of the first saltwater crocodilepox virus genome sequences from the world's largest living member of the Crocodylia.Sci Rep. 2018 Apr 4;8(1):5623. doi: 10.1038/s41598-018-23955-6. Sci Rep. 2018. PMID: 29618766 Free PMC article.
-
The genome of fowlpox virus.J Virol. 2000 Apr;74(8):3815-31. doi: 10.1128/jvi.74.8.3815-3831.2000. J Virol. 2000. PMID: 10729156 Free PMC article.
-
Crocodilepox Virus Evolutionary Genomics Supports Observed Poxvirus Infection Dynamics on Saltwater Crocodile (Crocodylus porosus).Viruses. 2019 Dec 2;11(12):1116. doi: 10.3390/v11121116. Viruses. 2019. PMID: 31810339 Free PMC article.
-
Evolution of viral DNA-dependent RNA polymerases.Virus Genes. 1995;11(2-3):271-84. doi: 10.1007/BF01728665. Virus Genes. 1995. PMID: 8828152 Review.
-
Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences.Crit Rev Biochem Mol Biol. 1993;28(5):375-430. doi: 10.3109/10409239309078440. Crit Rev Biochem Mol Biol. 1993. PMID: 8269709 Review.
Cited by
-
Prediction of a novel RNA binding domain in crocodilepox Zimbabwe Gene 157.Microb Inform Exp. 2011 Nov 21;1(1):12. doi: 10.1186/2042-5783-1-12. Microb Inform Exp. 2011. PMID: 22587704 Free PMC article.
-
Poxviral ankyrin proteins.Viruses. 2015 Feb 16;7(2):709-38. doi: 10.3390/v7020709. Viruses. 2015. PMID: 25690795 Free PMC article. Review.
-
Vaccinia virus F16 protein, a predicted catalytically inactive member of the prokaryotic serine recombinase superfamily, is targeted to nucleoli.Virology. 2011 Sep 1;417(2):334-42. doi: 10.1016/j.virol.2011.06.017. Epub 2011 Jul 12. Virology. 2011. PMID: 21752417 Free PMC article.
-
Poxviruses in bats … so what?Viruses. 2014 Apr 3;6(4):1564-77. doi: 10.3390/v6041564. Viruses. 2014. PMID: 24704730 Free PMC article. Review.
-
Molecular and microscopic characterization of a novel Eastern grey kangaroopox virus genome directly from a clinical sample.Sci Rep. 2017 Nov 28;7(1):16472. doi: 10.1038/s41598-017-16775-7. Sci Rep. 2017. Retraction in: Sci Rep. 2024 Aug 19;14(1):19153. doi: 10.1038/s41598-024-69649-0. PMID: 29184134 Free PMC article. Retracted.
References
-
- Abascal, F., R. Zardoya, and D. Posada. 2005. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21:2104-2105. - PubMed
-
- Afonso, C. L., E. R. Tulman, Z. Lu, L. Zsak, N. T. Sandybaev, U. Z. Kerembekova, V. L. Zaitsev, G. F. Kutish, and D. L. Rock. 2002. The genome of camelpox virus. Virology 295:1-9. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

