Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex
- PMID: 16625188
- PMCID: PMC5703407
- DOI: 10.1038/nature04716
Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex
Abstract
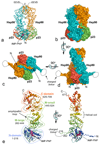
Hsp90 (heat shock protein of 90 kDa) is a ubiquitous molecular chaperone responsible for the assembly and regulation of many eukaryotic signalling systems and is an emerging target for rational chemotherapy of many cancers. Although the structures of isolated domains of Hsp90 have been determined, the arrangement and ATP-dependent dynamics of these in the full Hsp90 dimer have been elusive and contentious. Here we present the crystal structure of full-length yeast Hsp90 in complex with an ATP analogue and the co-chaperone p23/Sba1. The structure reveals the complex architecture of the 'closed' state of the Hsp90 chaperone, the extensive interactions between domains and between protein chains, the detailed conformational changes in the amino-terminal domain that accompany ATP binding, and the structural basis for stabilization of the closed state by p23/Sba1. Contrary to expectations, the closed Hsp90 would not enclose its client proteins but provides a bipartite binding surface whose formation and disruption are coupled to the chaperone ATPase cycle.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



Similar articles
-
Monitoring the Conformation of the Sba1/Hsp90 Complex in the Presence of Nucleotides with Mn(II)-Based Double Electron-Electron Resonance.J Phys Chem Lett. 2021 Dec 30;12(51):12235-12241. doi: 10.1021/acs.jpclett.1c03641. Epub 2021 Dec 20. J Phys Chem Lett. 2021. PMID: 34928609 Free PMC article.
-
Co-chaperone regulation of conformational switching in the Hsp90 ATPase cycle.J Biol Chem. 2004 Dec 10;279(50):51989-98. doi: 10.1074/jbc.M410562200. Epub 2004 Oct 2. J Biol Chem. 2004. PMID: 15466438
-
The Co-chaperone Sba1 connects the ATPase reaction of Hsp90 to the progression of the chaperone cycle.J Mol Biol. 2004 Oct 1;342(5):1403-13. doi: 10.1016/j.jmb.2004.07.064. J Mol Biol. 2004. PMID: 15364569
-
The Hsp90 chaperone machinery: conformational dynamics and regulation by co-chaperones.Biochim Biophys Acta. 2012 Mar;1823(3):624-35. doi: 10.1016/j.bbamcr.2011.09.003. Epub 2011 Sep 16. Biochim Biophys Acta. 2012. PMID: 21951723 Review.
-
Structure and mechanism of the Hsp90 molecular chaperone machinery.Annu Rev Biochem. 2006;75:271-94. doi: 10.1146/annurev.biochem.75.103004.142738. Annu Rev Biochem. 2006. PMID: 16756493 Review.
Cited by
-
Targeting Hsp90 in urothelial carcinoma.Oncotarget. 2015 Apr 20;6(11):8454-73. doi: 10.18632/oncotarget.3502. Oncotarget. 2015. PMID: 25909217 Free PMC article. Review.
-
Contributions of co-chaperones and post-translational modifications towards Hsp90 drug sensitivity.Future Med Chem. 2013 Jun;5(9):1059-71. doi: 10.4155/fmc.13.88. Future Med Chem. 2013. PMID: 23734688 Free PMC article. Review.
-
Dynamics of the regulation of Hsp90 by the co-chaperone Sti1.EMBO J. 2012 Mar 21;31(6):1518-28. doi: 10.1038/emboj.2012.37. Epub 2012 Feb 21. EMBO J. 2012. PMID: 22354036 Free PMC article.
-
Curcumin derivative C212 inhibits Hsp90 and eliminates both growing and quiescent leukemia cells in deep dormancy.Cell Commun Signal. 2020 Sep 29;18(1):159. doi: 10.1186/s12964-020-00652-4. Cell Commun Signal. 2020. PMID: 32993709 Free PMC article.
-
Substrate recognition and function of the R2TP complex in response to cellular stress.Front Genet. 2015 Feb 25;6:69. doi: 10.3389/fgene.2015.00069. eCollection 2015. Front Genet. 2015. PMID: 25767478 Free PMC article. Review.
References
-
- Mimnaugh EG, Chavany C, Neckers L. Polyubiquitination and proteasomal degradation of the p185c-erbB-2 receptor protein-tyrosine kinase induced by geldanamycin. J Biol Chem. 1996;271:22796–801. - PubMed
-
- Pearl LH. Hsp90 and Cdc37 -- a chaperone cancer conspiracy. Curr Opin Genet Dev. 2005;15:55–61. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

