CTL epitope distribution patterns in the Gag and Nef proteins of HIV-1 from subtype A infected subjects in Kenya: use of multiple peptide sets increases the detectable breadth of the CTL response
- PMID: 16620386
- PMCID: PMC1464141
- DOI: 10.1186/1471-2172-7-8
CTL epitope distribution patterns in the Gag and Nef proteins of HIV-1 from subtype A infected subjects in Kenya: use of multiple peptide sets increases the detectable breadth of the CTL response
Abstract
Background: Subtype A is a major strain in the HIV-1 pandemic in eastern Europe, central Asia and in certain regions of east Africa, notably in rural Kenya. While considerable effort has been focused upon mapping and defining immunodominant CTL epitopes in HIV-1 subtype B and subtype C infections, few epitope mapping studies have focused upon subtype A.
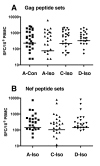
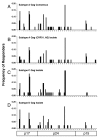
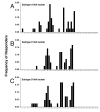
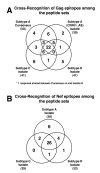
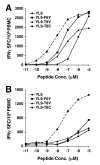
Results: We have used the IFN-gamma ELIspot assay and overlapping peptide pools to show that the pattern of CTL recognition of the Gag and Nef proteins in subtype A infection is similar to that seen in subtypes B and C. The p17 and p24 proteins of Gag and the central conserved region of Nef were targeted by CTL from HIV-1-infected Kenyans. Several epitope/HLA associations commonly seen in subtype B and C infection were also observed in subtype A infections. Notably, an immunodominant HLA-C restricted epitope (Gag 296-304; YL9) was observed, with 8/9 HLA-CW0304 subjects responding to this epitope. Screening the cohort with peptide sets representing subtypes A, C and D (the three most prevalent HIV-1 subtypes in east Africa), revealed that peptide sets based upon an homologous subtype (either isolate or consensus) only marginally improved the capacity to detect CTL responses. While the different peptide sets detected a similar number of responses (particularly in the Gag protein), each set was capable of detecting unique responses not identified with the other peptide sets.
Conclusion: Hence, screening with multiple peptide sets representing different sequences, and by extension different epitope variants, can increase the detectable breadth of the HIV-1-specific CTL response. Interpreting the true extent of cross-reactivity may be hampered by the use of 15-mer peptides at a single concentration and a lack of knowledge of the sequence that primed any given CTL response. Therefore, reagent choice and knowledge of the exact sequences that prime CTL responses will be important factors in experimentally defining cross-reactive CTL responses and their role in HIV-1 disease pathogenesis and validating vaccines aimed at generating broadly cross-reactive CTL responses.
Figures


Similar articles
-
Cytolytic T lymphocytes (CTLs) from HIV-1 subtype C-infected Indian patients recognize CTL epitopes from a conserved immunodominant region of HIV-1 Gag and Nef.J Infect Dis. 2005 Sep 1;192(5):749-59. doi: 10.1086/432547. Epub 2005 Jul 27. J Infect Dis. 2005. PMID: 16088824
-
Comprehensive screening for human immunodeficiency virus type 1 subtype-specific CD8 cytotoxic T lymphocytes and definition of degenerate epitopes restricted by HLA-A0207 and -C(W)0304 alleles.J Virol. 2002 May;76(10):4971-86. doi: 10.1128/jvi.76.10.4971-4986.2002. J Virol. 2002. PMID: 11967314 Free PMC article.
-
Differential narrow focusing of immunodominant human immunodeficiency virus gag-specific cytotoxic T-lymphocyte responses in infected African and caucasoid adults and children.J Virol. 2000 Jun;74(12):5679-90. doi: 10.1128/jvi.74.12.5679-5690.2000. J Virol. 2000. PMID: 10823876 Free PMC article.
-
Characterization of the Protective HIV-1 CTL Epitopes and the Corresponding HLA Class I Alleles: A Step towards Designing CTL Based HIV-1 Vaccine.Adv Virol. 2014;2014:321974. doi: 10.1155/2014/321974. Epub 2014 Mar 18. Adv Virol. 2014. PMID: 24744786 Free PMC article. Review.
-
The emerging role of HLA-C in HIV-1 infection.Immunology. 2011 Oct;134(2):116-22. doi: 10.1111/j.1365-2567.2011.03474.x. Immunology. 2011. PMID: 21896007 Free PMC article. Review.
Cited by
-
Group M consensus Gag and Nef peptides are as efficient at detecting clade A1 and D cross-subtype T-cell functions as subtype-specific consensus peptides.Vaccine. 2014 Jun 24;32(30):3787-95. doi: 10.1016/j.vaccine.2014.05.021. Epub 2014 May 14. Vaccine. 2014. PMID: 24837770 Free PMC article.
-
Use of RT-defective HIV virions: new tool to evaluate specific response in chronic asymptomatic HIV-infected individuals.PLoS One. 2013;8(3):e58927. doi: 10.1371/journal.pone.0058927. Epub 2013 Mar 14. PLoS One. 2013. PMID: 23516578 Free PMC article.
-
Nef modulates the immunogenicity of Gag encoded in a non-infectious HIV DNA vaccine.Vaccine. 2008 Jul 23;26(31):3795-804. doi: 10.1016/j.vaccine.2008.05.057. Epub 2008 Jun 17. Vaccine. 2008. PMID: 18586360 Free PMC article.
-
Multiple T-cell responses to human immunodeficiency virus type 1 are enhanced by dendritic cells.Clin Vaccine Immunol. 2009 Oct;16(10):1504-16. doi: 10.1128/CVI.00104-09. Epub 2009 Aug 19. Clin Vaccine Immunol. 2009. PMID: 19692626 Free PMC article.
-
Short communication: HIV-1 Nef protein carries multiple epitopes suitable for induction of cellular immunity for an HIV vaccine in Africa.AIDS Res Hum Retroviruses. 2014 Nov;30(11):1065-71. doi: 10.1089/AID.2013.0299. Epub 2014 Jul 16. AIDS Res Hum Retroviruses. 2014. PMID: 24866397 Free PMC article.
References
-
- Klausner RD, Fauci AS, Corey L, Nabel GJ, Gayle H, Berkley S, Haynes BF, Baltimore D, Collins C, Douglas RG, Esparza J, Francis DP, Ganguly NK, Gerberding JL, Johnston MI, Kazatchkine MD, McMichael AJ, Makgoba MW, Pantaleo G, Piot P, Shao Y, Tramont E, Varmus H, Wasserheit JN. Medicine. The need for a global HIV vaccine enterprise. Science. 2003;300:2036–2039. doi: 10.1126/science.1086916. - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

