The highly conserved family of Tetrahymena thermophila chromosome breakage elements contains an invariant 10-base-pair core
- PMID: 16607024
- PMCID: PMC1459666
- DOI: 10.1128/EC.5.4.771-780.2006
The highly conserved family of Tetrahymena thermophila chromosome breakage elements contains an invariant 10-base-pair core
Abstract
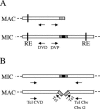
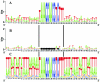
As a typical ciliate, Tetrahymena thermophila is a unicellular eukaryote that exhibits nuclear dimorphism: each cell contains a diploid, germ line micronucleus (MICN) and a polyploid, somatic macronucleus (MACN). During conjugation, when a new MACN differentiates from a mitotic descendant of the diploid fertilization nucleus, the five MICN chromosomes are site-specifically fragmented into 250 to 300 MACN chromosomes. The classic chromosome breakage sequence (CBS) is a 15-bp element (TAAACCAACCTCTTT) reported to be necessary and sufficient for chromosome breakage. To determine whether a CBS is present at every site of chromosome fragmentation and to assess the range of sequence variation tolerated, 31 CBSs were isolated without preconception as to the sequence of the chromosome breakage element. Additional CBS-related sequences were identified in the whole-genome sequence by their similarities to the classic CBS. Forty CBS elements behaved as authentic chromosome breakage sites. The CBS nucleotide sequence is more diverse than previously thought: nearly half of the CBS elements identified by unbiased methods have a variant of the classic CBS. Only an internal 10-bp core is completely conserved, but the entire 15-bp chromosome breakage sequence shows significant sequence conservation. Our results suggest that any one member of the CBS family provides a necessary and sufficient cis element for chromosome breakage. No chromosome breakage element totally unrelated to the classic CBS element was found; such elements, if they exist at all, must be rare.
Figures
Similar articles
-
Genome-wide characterization of tetrahymena thermophila chromosome breakage sites. I. Cloning and identification of functional sites.Genetics. 2005 Aug;170(4):1611-21. doi: 10.1534/genetics.104.031401. Epub 2005 Jun 14. Genetics. 2005. PMID: 15956677 Free PMC article.
-
Genome-wide characterization of Tetrahymena thermophila chromosome breakage sites. II. Physical and genetic mapping.Genetics. 2005 Aug;170(4):1623-31. doi: 10.1534/genetics.104.031435. Epub 2005 Jun 14. Genetics. 2005. PMID: 15956676 Free PMC article.
-
A long stringent sequence signal for programmed chromosome breakage in Tetrahymena thermophila.Nucleic Acids Res. 2000 Feb 15;28(4):895-900. doi: 10.1093/nar/28.4.895. Nucleic Acids Res. 2000. PMID: 10648780 Free PMC article.
-
Genome downsizing during ciliate development: nuclear division of labor through chromosome restructuring.Annu Rev Genet. 1996;30:557-78. doi: 10.1146/annurev.genet.30.1.557. Annu Rev Genet. 1996. PMID: 8982465 Review.
-
Mapping the germ-line and somatic genomes of a ciliated protozoan, Tetrahymena thermophila.Genome Res. 1998 Feb;8(2):91-9. doi: 10.1101/gr.8.2.91. Genome Res. 1998. PMID: 9477337 Review.
Cited by
-
Programmed Minichromosome Elimination as a Mechanism for Somatic Genome Reduction in Tetrahymena thermophila.PLoS Genet. 2016 Nov 2;12(11):e1006403. doi: 10.1371/journal.pgen.1006403. eCollection 2016 Nov. PLoS Genet. 2016. PMID: 27806059 Free PMC article.
-
Programmed DNA elimination in Tetrahymena: a small RNA-mediated genome surveillance mechanism.Adv Exp Med Biol. 2011;722:156-73. doi: 10.1007/978-1-4614-0332-6_10. Adv Exp Med Biol. 2011. PMID: 21915788 Free PMC article. Review.
-
Tetrahymena thermophila, a unicellular eukaryote with separate germline and somatic genomes.Res Microbiol. 2011 Jul-Aug;162(6):578-86. doi: 10.1016/j.resmic.2011.05.001. Epub 2011 May 18. Res Microbiol. 2011. PMID: 21624459 Free PMC article. Review.
-
Programmed chromosome fragmentation in ciliated protozoa: multiple means to chromosome ends.Microbiol Mol Biol Rev. 2023 Dec 20;87(4):e0018422. doi: 10.1128/mmbr.00184-22. Epub 2023 Nov 27. Microbiol Mol Biol Rev. 2023. PMID: 38009915 Free PMC article. Review.
-
Use of HAPPY mapping for the higher order assembly of the Tetrahymena genome.Genomics. 2006 Oct;88(4):443-51. doi: 10.1016/j.ygeno.2006.05.002. Epub 2006 Jun 19. Genomics. 2006. PMID: 16782302 Free PMC article.
References
-
- Bailey, T. L., and C. Elkan. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2:28-36. - PubMed
-
- Bailey, T. L., and M. Gribskov. 1998. Combining evidence using p-values: application to sequence homology searches. Bioinformatics 14:48-54. - PubMed
-
- Blackburn, E. H., and J. C. Gall. 1978. A tandemly repeated sequence at the termini of the extrachromosomal ribosomal RNA genes in Tetrahymena. J. Mol. Biol. 120:33-53. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

