Phylogenetic analyses of Vitis (Vitaceae) based on complete chloroplast genome sequences: effects of taxon sampling and phylogenetic methods on resolving relationships among rosids
- PMID: 16603088
- PMCID: PMC1479384
- DOI: 10.1186/1471-2148-6-32
Phylogenetic analyses of Vitis (Vitaceae) based on complete chloroplast genome sequences: effects of taxon sampling and phylogenetic methods on resolving relationships among rosids
Abstract
Background: The Vitaceae (grape) is an economically important family of angiosperms whose phylogenetic placement is currently unresolved. Recent phylogenetic analyses based on one to several genes have suggested several alternative placements of this family, including sister to Caryophyllales, asterids, Saxifragales, Dilleniaceae or to rest of rosids, though support for these different results has been weak. There has been a recent interest in using complete chloroplast genome sequences for resolving phylogenetic relationships among angiosperms. These studies have clarified relationships among several major lineages but they have also emphasized the importance of taxon sampling and the effects of different phylogenetic methods for obtaining accurate phylogenies. We sequenced the complete chloroplast genome of Vitis vinifera and used these data to assess relationships among 27 angiosperms, including nine taxa of rosids.
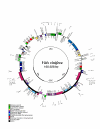
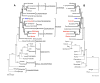
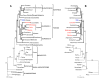
Results: The Vitis vinifera chloroplast genome is 160,928 bp in length, including a pair of inverted repeats of 26,358 bp that are separated by small and large single copy regions of 19,065 bp and 89,147 bp, respectively. The gene content and order of Vitis is identical to many other unrearranged angiosperm chloroplast genomes, including tobacco. Phylogenetic analyses using maximum parsimony and maximum likelihood were performed on DNA sequences of 61 protein-coding genes for two datasets with 28 or 29 taxa, including eight or nine taxa from four of the seven currently recognized major clades of rosids. Parsimony and likelihood phylogenies of both data sets provide strong support for the placement of Vitaceae as sister to the remaining rosids. However, the position of the Myrtales and support for the monophyly of the eurosid I clade differs between the two data sets and the two methods of analysis. In parsimony analyses, the inclusion of Gossypium is necessary to obtain trees that support the monophyly of the eurosid I clade. However, maximum likelihood analyses place Cucumis as sister to the Myrtales and therefore do not support the monophyly of the eurosid I clade.
Conclusion: Phylogenies based on DNA sequences from complete chloroplast genome sequences provide strong support for the position of the Vitaceae as the earliest diverging lineage of rosids. Our phylogenetic analyses support recent assertions that inadequate taxon sampling and incorrect model specification for concatenated multi-gene data sets can mislead phylogenetic inferences when using whole chloroplast genomes for phylogeny reconstruction.
Figures
Similar articles
-
The complete chloroplast genome sequence of Citrus sinensis (L.) Osbeck var 'Ridge Pineapple': organization and phylogenetic relationships to other angiosperms.BMC Plant Biol. 2006 Sep 30;6:21. doi: 10.1186/1471-2229-6-21. BMC Plant Biol. 2006. PMID: 17010212 Free PMC article.
-
The Complete Chloroplast Genome Sequence of Ampelopsis: Gene Organization, Comparative Analysis, and Phylogenetic Relationships to Other Angiosperms.Front Plant Sci. 2016 Mar 21;7:341. doi: 10.3389/fpls.2016.00341. eCollection 2016. Front Plant Sci. 2016. PMID: 27047519 Free PMC article.
-
The complete chloroplast genome sequence of Gossypium hirsutum: organization and phylogenetic relationships to other angiosperms.BMC Genomics. 2006 Mar 23;7:61. doi: 10.1186/1471-2164-7-61. BMC Genomics. 2006. PMID: 16553962 Free PMC article.
-
Nuclear phylogenomics of angiosperms and insights into their relationships and evolution.J Integr Plant Biol. 2024 Mar;66(3):546-578. doi: 10.1111/jipb.13609. Epub 2024 Jan 30. J Integr Plant Biol. 2024. PMID: 38289011 Review.
-
The impact of taxon sampling on phylogenetic inference: a review of two decades of controversy.Brief Bioinform. 2012 Jan;13(1):122-34. doi: 10.1093/bib/bbr014. Epub 2011 Mar 23. Brief Bioinform. 2012. PMID: 21436145 Free PMC article. Review.
Cited by
-
The complete chloroplast genome sequence of Citrus sinensis (L.) Osbeck var 'Ridge Pineapple': organization and phylogenetic relationships to other angiosperms.BMC Plant Biol. 2006 Sep 30;6:21. doi: 10.1186/1471-2229-6-21. BMC Plant Biol. 2006. PMID: 17010212 Free PMC article.
-
Complete plastome sequences of Equisetum arvense and Isoetes flaccida: implications for phylogeny and plastid genome evolution of early land plant lineages.BMC Evol Biol. 2010 Oct 23;10:321. doi: 10.1186/1471-2148-10-321. BMC Evol Biol. 2010. PMID: 20969798 Free PMC article.
-
Using phylogenomic patterns and gene ontology to identify proteins of importance in plant evolution.Genome Biol Evol. 2010 Jul 12;2:225-39. doi: 10.1093/gbe/evq012. Genome Biol Evol. 2010. PMID: 20624728 Free PMC article.
-
Comparative chloroplast genomics and phylogenetics of Fagopyrum esculentum ssp. ancestrale -a wild ancestor of cultivated buckwheat.BMC Plant Biol. 2008 May 20;8:59. doi: 10.1186/1471-2229-8-59. BMC Plant Biol. 2008. PMID: 18492277 Free PMC article.
-
Sequencing of chloroplast genomes from wheat, barley, rye and their relatives provides a detailed insight into the evolution of the Triticeae tribe.PLoS One. 2014 Mar 10;9(3):e85761. doi: 10.1371/journal.pone.0085761. eCollection 2014. PLoS One. 2014. PMID: 24614886 Free PMC article.
References
-
- Soltis DE, Soltis PS, Endress PK, Chase MW. Phylogeny and evolution of Angiosperms. Sunderland Massachusetts: Sinauer Associates Inc; 2005.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

