Cap-independent translation mechanism of red clover necrotic mosaic virus RNA2 differs from that of RNA1 and is linked to RNA replication
- PMID: 16571795
- PMCID: PMC1440462
- DOI: 10.1128/JVI.80.8.3781-3791.2006
Cap-independent translation mechanism of red clover necrotic mosaic virus RNA2 differs from that of RNA1 and is linked to RNA replication
Abstract
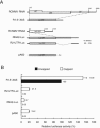
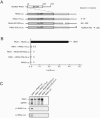
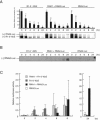
The genome of Red clover necrotic mosaic virus (RCNMV) in the genus Dianthovirus is divided into two RNA molecules of RNA1 and RNA2, which have no cap structure at the 5' end and no poly(A) tail at the 3' end. The 3' untranslated region (3' UTR) of RCNMV RNA1 contains an essential RNA element (3'TE-DR1), which is required for cap-independent translation. In this study, we investigated a cap-independent translational mechanism of RNA2 using a firefly luciferase (Luc) gene expression assay system in cowpea protoplasts and a cell-free lysate (BYL) prepared from evacuolated tobacco BY2 protoplasts. We were unable to detect cis-acting RNA sequences in RNA2 that can replace the function of a cap structure, such as the 3'TE-DR1 of RNA1. However, the uncapped reporter RNA2, RNA2-Luc, in which the Luc open reading frame (ORF) was inserted between the 5' UTR and the movement protein ORF, was effectively translated in the presence of p27 and p88 in protoplasts in which RNA2-Luc was replicated. Time course experiments in protoplasts showed that the translational activity of RNA2-Luc did not reflect the amount of RNA2. Mutations in cis-acting RNA replication elements of RNA2 abolished the cap-independent translational activity of RNA2-Luc, suggesting that the translational activity of RNA2-Luc is coupled to RNA replication. Our results show that the translational mechanism differs between two segmented genomic RNAs of RCNMV. We present a model in which only RNA2 that is generated de novo through the viral RNA replication machinery functions as mRNA for translation.
Figures



Similar articles
-
Cap-independent translational enhancement by the 3' untranslated region of red clover necrotic mosaic virus RNA1.J Virol. 2003 Nov;77(22):12113-21. doi: 10.1128/jvi.77.22.12113-12121.2003. J Virol. 2003. PMID: 14581548 Free PMC article.
-
Host-dependent roles of the viral 5' untranslated region (UTR) in RNA stabilization and cap-independent translational enhancement mediated by the 3' UTR of Red clover necrotic mosaic virus RNA1.Virology. 2009 Aug 15;391(1):107-18. doi: 10.1016/j.virol.2009.05.037. Epub 2009 Jul 4. Virology. 2009. PMID: 19577782
-
cis-Acting core RNA elements required for negative-strand RNA synthesis and cap-independent translation are separated in the 3'-untranslated region of Red clover necrotic mosaic virus RNA1.Virology. 2007 Dec 5;369(1):168-81. doi: 10.1016/j.virol.2007.07.016. Epub 2007 Aug 28. Virology. 2007. PMID: 17727911
-
Cap-independent translation of plant viral RNAs.Virus Res. 2006 Jul;119(1):63-75. doi: 10.1016/j.virusres.2005.10.010. Epub 2005 Dec 19. Virus Res. 2006. PMID: 16360925 Free PMC article. Review.
-
Molecular biology and epidemiology of dianthoviruses.Adv Virus Res. 2013;87:37-74. doi: 10.1016/B978-0-12-407698-3.00002-8. Adv Virus Res. 2013. PMID: 23809920 Free PMC article. Review.
Cited by
-
A 3'-end structure in RNA2 of a crinivirus is essential for viral RNA synthesis and contributes to replication-associated translation activity.Sci Rep. 2016 Oct 3;6:34482. doi: 10.1038/srep34482. Sci Rep. 2016. PMID: 27694962 Free PMC article.
-
Differential roles of Hsp70 and Hsp90 in the assembly of the replicase complex of a positive-strand RNA plant virus.J Virol. 2012 Nov;86(22):12091-104. doi: 10.1128/JVI.01659-12. Epub 2012 Aug 29. J Virol. 2012. PMID: 22933272 Free PMC article.
-
Replication-coupled packaging mechanism in positive-strand RNA viruses: synchronized coexpression of functional multigenome RNA components of an animal and a plant virus in Nicotiana benthamiana cells by agroinfiltration.J Virol. 2008 Feb;82(3):1484-95. doi: 10.1128/JVI.01540-07. Epub 2007 Nov 21. J Virol. 2008. PMID: 18032497 Free PMC article.
-
A trans-activator-like structure in RCNMV RNA1 evokes the origin of the trans-activator in RNA2.PLoS Pathog. 2020 Jan 6;16(1):e1008271. doi: 10.1371/journal.ppat.1008271. eCollection 2020 Jan. PLoS Pathog. 2020. PMID: 31905231 Free PMC article.
-
GAPDH--a recruits a plant virus movement protein to cortical virus replication complexes to facilitate viral cell-to-cell movement.PLoS Pathog. 2014 Nov 20;10(11):e1004505. doi: 10.1371/journal.ppat.1004505. eCollection 2014 Nov. PLoS Pathog. 2014. PMID: 25411849 Free PMC article.
References
-
- Blumenthal, T., and G. G. Carmichael. 1979. RNA replication: function and structure of Qβ-replicase. Annu. Rev. Biochem. 48:525-548. - PubMed
-
- Choi, S. B., C. Wang, D. G. Muench, K. Ozawa, V. R. Franceschi, Y. Wu, and T. W. Okita. 2000. Messenger RNA targeting of rice seed storage proteins to specific ER subdomains. Nature 407:765-767. - PubMed
-
- Damayanti, T. A., H. Nagano, K. Mise, I. Furusawa, and T. Okuno. 2002. Positional effect of deletions on viability, especially on encapsidation, of brome mosaic virus D-RNA in barley protoplasts. Virology 293:314-319. - PubMed
-
- Doi, N., S. Zenno, R. Ueda, H. Ohki-Hamazaki, K. Ui-Tei, and K. Saigo. 2003. Short-interfering-RNA-mediated gene silencing in mammalian cells requires Dicer and eIF2C translation initiation factors. Curr. Biol. 13:41-46. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

