The complete chloroplast genome sequence of Gossypium hirsutum: organization and phylogenetic relationships to other angiosperms
- PMID: 16553962
- PMCID: PMC1513215
- DOI: 10.1186/1471-2164-7-61
The complete chloroplast genome sequence of Gossypium hirsutum: organization and phylogenetic relationships to other angiosperms
Abstract
Background: Cotton (Gossypium hirsutum) is the most important fiber crop grown in 90 countries. In 2004-2005, US farmers planted 79% of the 5.7-million hectares of nuclear transgenic cotton. Unfortunately, genetically modified cotton has the potential to hybridize with other cultivated and wild relatives, resulting in geographical restrictions to cultivation. However, chloroplast genetic engineering offers the possibility of containment because of maternal inheritance of transgenes. The complete chloroplast genome of cotton provides essential information required for genetic engineering. In addition, the sequence data were used to assess phylogenetic relationships among the major clades of rosids using cotton and 25 other completely sequenced angiosperm chloroplast genomes.
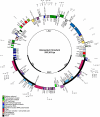
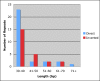
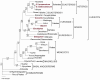
Results: The complete cotton chloroplast genome is 160,301 bp in length, with 112 unique genes and 19 duplicated genes within the IR, containing a total of 131 genes. There are four ribosomal RNAs, 30 distinct tRNA genes and 17 intron-containing genes. The gene order in cotton is identical to that of tobacco but lacks rpl22 and infA. There are 30 direct and 24 inverted repeats 30 bp or longer with a sequence identity > or = 90%. Most of the direct repeats are within intergenic spacer regions, introns and a 72 bp-long direct repeat is within the psaA and psaB genes. Comparison of protein coding sequences with expressed sequence tags (ESTs) revealed nucleotide substitutions resulting in amino acid changes in ndhC, rpl23, rpl20, rps3 and clpP. Phylogenetic analysis of a data set including 61 protein-coding genes using both maximum likelihood and maximum parsimony were performed for 28 taxa, including cotton and five other angiosperm chloroplast genomes that were not included in any previous phylogenies.
Conclusion: Cotton chloroplast genome lacks rpl22 and infA and contains a number of dispersed direct and inverted repeats. RNA editing resulted in amino acid changes with significant impact on their hydropathy. Phylogenetic analysis provides strong support for the position of cotton in the Malvales in the eurosids II clade sister to Arabidopsis in the Brassicales. Furthermore, there is strong support for the placement of the Myrtales sister to the eurosid I clade, although expanded taxon sampling is needed to further test this relationship.
Figures
Similar articles
-
The complete chloroplast genome sequence of Citrus sinensis (L.) Osbeck var 'Ridge Pineapple': organization and phylogenetic relationships to other angiosperms.BMC Plant Biol. 2006 Sep 30;6:21. doi: 10.1186/1471-2229-6-21. BMC Plant Biol. 2006. PMID: 17010212 Free PMC article.
-
Phylogenetic analyses of Vitis (Vitaceae) based on complete chloroplast genome sequences: effects of taxon sampling and phylogenetic methods on resolving relationships among rosids.BMC Evol Biol. 2006 Apr 9;6:32. doi: 10.1186/1471-2148-6-32. BMC Evol Biol. 2006. PMID: 16603088 Free PMC article.
-
Complete plastid genome sequence of Daucus carota: implications for biotechnology and phylogeny of angiosperms.BMC Genomics. 2006 Aug 31;7:222. doi: 10.1186/1471-2164-7-222. BMC Genomics. 2006. PMID: 16945140 Free PMC article.
-
The complete nucleotide sequence of the coffee (Coffea arabica L.) chloroplast genome: organization and implications for biotechnology and phylogenetic relationships amongst angiosperms.Plant Biotechnol J. 2007 Mar;5(2):339-53. doi: 10.1111/j.1467-7652.2007.00245.x. Plant Biotechnol J. 2007. PMID: 17309688 Free PMC article.
-
Green giant-a tiny chloroplast genome with mighty power to produce high-value proteins: history and phylogeny.Plant Biotechnol J. 2021 Mar;19(3):430-447. doi: 10.1111/pbi.13556. Epub 2021 Feb 22. Plant Biotechnol J. 2021. PMID: 33484606 Free PMC article. Review.
Cited by
-
Complete Arabis alpina chloroplast genome sequence and insight into its polymorphism.Meta Gene. 2013 Nov 15;1:65-75. doi: 10.1016/j.mgene.2013.10.004. eCollection 2013 Dec. Meta Gene. 2013. PMID: 25606376 Free PMC article.
-
Genetic, evolutionary and phylogenetic aspects of the plastome of annatto (Bixa orellana L.), the Amazonian commercial species of natural dyes.Planta. 2019 Feb;249(2):563-582. doi: 10.1007/s00425-018-3023-6. Epub 2018 Oct 11. Planta. 2019. PMID: 30310983
-
Castor bean organelle genome sequencing and worldwide genetic diversity analysis.PLoS One. 2011;6(7):e21743. doi: 10.1371/journal.pone.0021743. Epub 2011 Jul 7. PLoS One. 2011. PMID: 21750729 Free PMC article.
-
The newly assembled chloroplast genome of Aeluropus littoralis: molecular feature characterization and phylogenetic analysis with related species.Sci Rep. 2024 Mar 18;14(1):6472. doi: 10.1038/s41598-024-57141-8. Sci Rep. 2024. PMID: 38499663 Free PMC article.
-
Methods of analysis of chloroplast genomes of C3, Kranz type C4 and Single Cell C4 photosynthetic members of Chenopodiaceae.Plant Methods. 2020 Aug 31;16:119. doi: 10.1186/s13007-020-00662-w. eCollection 2020. Plant Methods. 2020. PMID: 32874195 Free PMC article.
References
-
- Palmer JD. Plastid chromosomes: structure and evolution. In: Bogorad L, Vasil K, editor. The Molecular Biology of Plastids. San Diego: Academic Press; 1991. pp. 5–53.
-
- Raubeson LA, Jansen RK. Chloroplast genomes of plants. In: Henry H, editor. Diversity and Evolution of Plants-Genotypic and Phenotypic Variation in Higher Plants. Wallingford: CABI Publishing; 2005. pp. 45–68.
-
- Saski C, Lee S, Daniell H, Wood T, Tomkins J, Kim H-G, Jansen RK. Complete chloroplast genome sequence of Glycine max and comparative analyses with other legume genomes. Plt Mol Biol. 2005;59:309–322. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

