A tripartite DNA-binding element, comprised of the nuclear localization signal and two AT-hook motifs, mediates the association of LEDGF/p75 with chromatin in vivo
- PMID: 16549878
- PMCID: PMC1405818
- DOI: 10.1093/nar/gkl052
A tripartite DNA-binding element, comprised of the nuclear localization signal and two AT-hook motifs, mediates the association of LEDGF/p75 with chromatin in vivo
Abstract
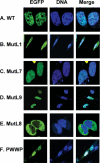
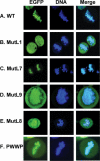
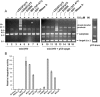
Lens epithelium-derived growth factor p75 (LEDGF/p75) is a DNA-binding, transcriptional co-activator that participates in HIV-1 integration site targeting. Using complementary approaches, we determined the mechanisms of LEDGF/p75 DNA-binding in vitro and chromatin-association in living cells. The binding of highly-purified, recombinant protein was assayed by surface plasmon resonance (SPR) and electrophoretic mobility gel shift. Neither assay revealed evidence for sequence-specific DNA-binding. Residues 146-197 spanning the nuclear localization signal (NLS) and two AT-hook motifs mediated non-specific DNA-binding, and DNA-binding deficient mutants retained the ability to efficiently stimulate HIV-1 integrase activity in vitro. Chromatin-association was assessed by visualizing the localization of EGFP fusion proteins in interphase and mitotic cells. Although a conserved N-terminal PWWP domain was not required for binding to condensed mitotic chromosomes, its deletion subtly affected the nucleoplasmic distribution of the protein during interphase. A dual AT-hook mutant associated normally with chromatin, yet when the mutations were combined with NLS changes or deletion of the PWWP domain, chromatin-binding function was lost. As the PWWP domain did not readily bind free DNA in vitro, our results indicate that chromatin-association is primarily affected through DNA-binding, with the PWWP domain likely contributing a protein interaction to the overall affinity of LEDGF/p75 for human chromatin.
Figures



Similar articles
-
Identification of the LEDGF/p75 HIV-1 integrase-interaction domain and NLS reveals NLS-independent chromatin tethering.J Cell Sci. 2005 Apr 15;118(Pt 8):1733-43. doi: 10.1242/jcs.02299. Epub 2005 Mar 29. J Cell Sci. 2005. PMID: 15797927
-
Identification and characterization of the chromatin-binding domains of the HIV-1 integrase interactor LEDGF/p75.J Mol Biol. 2006 Jul 21;360(4):760-73. doi: 10.1016/j.jmb.2006.04.073. Epub 2006 May 17. J Mol Biol. 2006. PMID: 16793062
-
Role of the PWWP domain of lens epithelium-derived growth factor (LEDGF)/p75 cofactor in lentiviral integration targeting.J Biol Chem. 2011 Dec 2;286(48):41812-41826. doi: 10.1074/jbc.M111.255711. Epub 2011 Oct 10. J Biol Chem. 2011. Retraction in: J Biol Chem. 2018 Jan 5;293(1):114. doi: 10.1074/jbc.W117.001458. PMID: 21987578 Free PMC article. Retracted.
-
Protein-protein and protein-chromatin interactions of LEDGF/p75 as novel drug targets.Drug Discov Today Technol. 2017 Jun;24:25-31. doi: 10.1016/j.ddtec.2017.11.002. Epub 2017 Nov 23. Drug Discov Today Technol. 2017. PMID: 29233296 Review.
-
Host factors for retroviral integration site selection.Trends Biochem Sci. 2015 Feb;40(2):108-16. doi: 10.1016/j.tibs.2014.12.001. Epub 2014 Dec 31. Trends Biochem Sci. 2015. PMID: 25555456 Review.
Cited by
-
Identification and functional analysis of NOL7 nuclear and nucleolar localization signals.BMC Cell Biol. 2010 Sep 27;11:74. doi: 10.1186/1471-2121-11-74. BMC Cell Biol. 2010. PMID: 20875127 Free PMC article.
-
Nuclear protein LEDGF/p75 recognizes supercoiled DNA by a novel DNA-binding domain.Nucleic Acids Res. 2011 Jul;39(12):5067-81. doi: 10.1093/nar/gkr088. Epub 2011 Feb 23. Nucleic Acids Res. 2011. PMID: 21345933 Free PMC article.
-
HIV-1 integrase modulates the interaction of the HIV-1 cellular cofactor LEDGF/p75 with chromatin.Retrovirology. 2011 Apr 21;8:27. doi: 10.1186/1742-4690-8-27. Retrovirology. 2011. PMID: 21510906 Free PMC article.
-
Bimodal high-affinity association of Brd4 with murine leukemia virus integrase and mononucleosomes.Nucleic Acids Res. 2014 Apr;42(8):4868-81. doi: 10.1093/nar/gku135. Epub 2014 Feb 11. Nucleic Acids Res. 2014. PMID: 24520112 Free PMC article.
-
LEDGF/p75-independent HIV-1 replication demonstrates a role for HRP-2 and remains sensitive to inhibition by LEDGINs.PLoS Pathog. 2012;8(3):e1002558. doi: 10.1371/journal.ppat.1002558. Epub 2012 Mar 1. PLoS Pathog. 2012. PMID: 22396646 Free PMC article.
References
-
- Spiegelman B.M., Heinrich R. Biological control through regulated transcriptional coactivators. Cell. 2004;119:157–167. - PubMed
-
- Shinohara T., Singh D.P., Fatma N. LEDGF, a survival factor, activates stress-related genes. Prog. Retin. Eye Res. 2002;21:341–358. - PubMed
-
- Wu X., Daniels T., Molinaro C., Lilly M.B., Casiano C.A. Caspase cleavage of the nuclear autoantigen LEDGF/p75 abrogates its pro-survival function: implications for autoimmunity in atopic disorders. Cell Death Differ. 2002;9:915–925. - PubMed
-
- Daniels T., Zhang J., Gutierrez I., Elliot M.L., Yamada B., Heeb M.J., Sheets S.M., Wu X., Casiano C.A. Antinuclear autoantibodies in prostate cancer: immunity to LEDGF/p75, a survival protein highly expressed in prostate tumors and cleaved during apoptosis. Prostate. 2005;62:14–26. - PubMed

