DNA deformability changes of single base pair mutants within CDE binding sites in S. Cerevisiae centromere DNA correlate with measured chromosomal loss rates and CDE binding site symmetries
- PMID: 16542422
- PMCID: PMC1434758
- DOI: 10.1186/1471-2199-7-12
DNA deformability changes of single base pair mutants within CDE binding sites in S. Cerevisiae centromere DNA correlate with measured chromosomal loss rates and CDE binding site symmetries
Abstract
Background: The centromeres in yeast (S. cerevisiae) are organized by short DNA sequences (125 bp) on each chromosome consisting of 2 conserved elements: CDEI and CDEIII spaced by a CDEII region. CDEI and CDEIII are critical sequence specific protein binding sites necessary for correct centromere formation and following assembly with proteins, are positioned near each other on a specialized nucleosome. Hegemann et al. BioEssays 1993, 15: 451-460 reported single base DNA mutants within the critical CDEI and CDEIII binding sites on the centromere of chromosome 6 and quantitated centromere loss of function, which they measured as loss rates for the different chromosome 6 mutants during cell division. Olson et al. Proc Natl Acad Sci USA 1998, 95: 11163-11168 reported the use of protein-DNA crystallography data to produce a DNA dinucleotide protein deformability energetic scale (PD-scale) that describes local DNA deformability by sequence specific binding proteins. We have used the PD-scale to investigate the DNA sequence dependence of the yeast chromosome 6 mutants' loss rate data. Each single base mutant changes 2 PD-scale values at that changed base position relative to the wild type. In this study, we have utilized these mutants to demonstrate a correlation between the change in DNA deformability of the CDEI and CDEIII core sites and the overall experimentally measured chromosome loss rates of the chromosome 6 mutants.
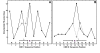
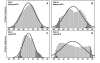
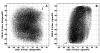
Results: In the CDE I and CDEIII core binding regions an increase in the magnitude of change in deformability of chromosome 6 single base mutants with respect to the wild type correlates to an increase in the measured chromosome loss rate. These correlations were found to be significant relative to 10(5) Monte Carlo randomizations of the dinucleotide PD-scale applied to the same calculation. A net loss of deformability also tends to increase the loss rate. Binding site position specific, 4 data-point correlations were also created using the wild type sequence and the 3 associated alternate base mutants at each binding site position. These position specific slope magnitudes, or sensitivities, correlated with and reflected the underlying position symmetry of the DNA binding sequences.
Conclusion: These results suggest the utility of correlating quantitative aspects of sequence specific protein-DNA complex single base mutants with changes in the easily calculated PD-deformability scale of the individual DNA sequence mutants. Using this PD approach, it may be possible in the future to understand the magnitude of biological or energetic functional effects of specific DNA sequence mutants within DNA-protein complexes in terms of their effect on DNA deformability.
Figures
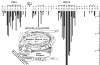

Similar articles
-
CSE4 genetically interacts with the Saccharomyces cerevisiae centromere DNA elements CDE I and CDE II but not CDE III. Implications for the path of the centromere dna around a cse4p variant nucleosome.Genetics. 2000 Nov;156(3):973-81. doi: 10.1093/genetics/156.3.973. Genetics. 2000. PMID: 11063678 Free PMC article.
-
Cpf1 protein induced bending of yeast centromere DNA element I.Nucleic Acids Res. 1993 Oct 11;21(20):4726-33. doi: 10.1093/nar/21.20.4726. Nucleic Acids Res. 1993. PMID: 8233820 Free PMC article.
-
The yeast centromere CDEI/Cpf1 complex: differences between in vitro binding and in vivo function.Nucleic Acids Res. 1994 Jul 25;22(14):2791-800. doi: 10.1093/nar/22.14.2791. Nucleic Acids Res. 1994. PMID: 8052535 Free PMC article.
-
The centromere of budding yeast.Bioessays. 1993 Jul;15(7):451-60. doi: 10.1002/bies.950150704. Bioessays. 1993. PMID: 8379948 Review.
-
The structure and function of yeast centromeres.Annu Rev Genet. 1985;19:29-55. doi: 10.1146/annurev.ge.19.120185.000333. Annu Rev Genet. 1985. PMID: 3909945 Review. No abstract available.
Cited by
-
RrS1-like sequences of water frogs from Central Europe and around the Aegean Sea: chromosomal organization, evolution, possible function.J Mol Evol. 2011 Apr;72(4):368-82. doi: 10.1007/s00239-011-9436-5. Epub 2011 Mar 20. J Mol Evol. 2011. PMID: 21424546
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

