Analyses of subgenomic promoters of Hibiscus chlorotic ringspot virus and demonstration of 5' untranslated region and 3'-terminal sequences functioning as subgenomic promoters
- PMID: 16537607
- PMCID: PMC1440410
- DOI: 10.1128/JVI.80.7.3395-3405.2006
Analyses of subgenomic promoters of Hibiscus chlorotic ringspot virus and demonstration of 5' untranslated region and 3'-terminal sequences functioning as subgenomic promoters
Abstract
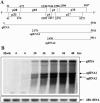
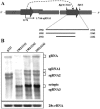
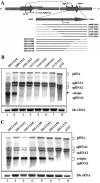
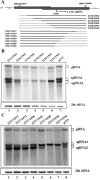
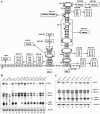
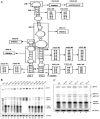
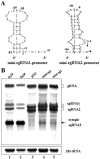
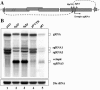
Hibiscus chlorotic ringspot virus (HCRSV), which belongs to the genus Carmovirus, generates two 3'-coterminal subgenomic RNAs (sgRNAs) of 1.4 kb and 1.7 kb. Transcription start sites of the two sgRNAs were identified at nucleotides (nt) 2178 and 2438, respectively. The full promoter of sgRNA1, a 118-base sequence, is localized between positions +6 and -112 relative to its transcription start site (+1). Similarly, a 132-base sequence, from +6 to -126, defines the sgRNA2 promoter. Computer analysis revealed that both sgRNA promoters share a similar two-stem-loop (SL1 + SL2) structure, immediately upstream of the transcription start site. Mutational analysis of the primary sequence and secondary structures showed further similarities between the two subgenomic promoters. The basal portion of SL2, encompassing the transcription start site, was essential for transcription activity in each promoter, while SL1 and the upper portion of SL2 played a role in transcription enhancement. Both the 5' untranslated region (UTR) and the last 87 nt at the 3' UTR of HCRSV genomic RNA are likely to be the putative genomic plus-strand and minus-strand promoters, respectively. They function well as individual sgRNA promoters to produce ectopic subgenomic RNAs in vivo but not to the same levels of the actual sgRNA promoters. This suggests that HCRSV sgRNA promoters share common features with the promoters for genomic plus-strand and minus-strand RNA synthesis. To our knowledge, this is the first demonstration that both the 5' UTR and part of the 3' UTR can be duplicated and function as sgRNA promoters within a single viral genome.
Figures

Similar articles
-
Mapping of the Tobacco mosaic virus movement protein and coat protein subgenomic RNA promoters in vivo.Virology. 2000 Sep 15;275(1):177-92. doi: 10.1006/viro.2000.0511. Virology. 2000. PMID: 11017798
-
Analysis of the two subgenomic RNA promoters for turnip crinkle virus in vivo and in vitro.Virology. 1997 May 26;232(1):174-86. doi: 10.1006/viro.1997.8550. Virology. 1997. PMID: 9185601
-
Significance of the 3'-terminal region in minus-strand RNA synthesis of Hibiscus chlorotic ringspot virus.J Gen Virol. 2004 Jun;85(Pt 6):1763-1776. doi: 10.1099/vir.0.79861-0. J Gen Virol. 2004. PMID: 15166462
-
[Structure and function of the non-coding regions of hepatitis C viral RNA].Postepy Biochem. 2006;52(1):62-71. Postepy Biochem. 2006. PMID: 16869303 Review. Polish.
-
[Structure and function of HAV genome (RNA)].Nihon Rinsho. 2004 Aug;62 Suppl 8:428-32. Nihon Rinsho. 2004. PMID: 15453360 Review. Japanese. No abstract available.
Cited by
-
Evidence for a premature termination mechanism of subgenomic mRNA transcription in a carmovirus.J Virol. 2010 Aug;84(15):7904-7. doi: 10.1128/JVI.00742-10. Epub 2010 May 26. J Virol. 2010. PMID: 20504939 Free PMC article.
-
Hibiscus chlorotic ringspot virus coat protein is essential for cell-to-cell and long-distance movement but not for viral RNA replication.PLoS One. 2014 Nov 17;9(11):e113347. doi: 10.1371/journal.pone.0113347. eCollection 2014. PLoS One. 2014. PMID: 25402344 Free PMC article.
-
The conserved stem-loop II structure at the 3' untranslated region of Japanese encephalitis virus genome is required for the formation of subgenomic flaviviral RNA.PLoS One. 2018 Jul 26;13(7):e0201250. doi: 10.1371/journal.pone.0201250. eCollection 2018. PLoS One. 2018. PMID: 30048535 Free PMC article.
-
Characterization of Hibiscus Chlorotic Ringspot Virus-Derived vsiRNAs from Infected Hibiscus rosa-sinensis in China.Plant Pathol J. 2024 Oct;40(5):415-424. doi: 10.5423/PPJ.OA.06.2024.0090. Epub 2024 Oct 1. Plant Pathol J. 2024. PMID: 39397297 Free PMC article.
References
-
- Adkins, S., and C. C. Kao. 1998. Subgenomic RNA promoters dictate the mode of recognition by bromoviral RNA-dependent RNA polymerases. Virology 252:1-8. - PubMed
-
- Balmori, E., D. Gilmer, K. Richards, H. Guilley, and G. Jonard. 1993. Mapping the promoter for subgenomic RNA synthesis on beet necrotic yellow vein virus RNA 3. Biochimie 75:517-521. - PubMed
-
- Grdzelishvili, V. Z., S. N. Chapman, W. O. Dawson, and D. J. Lewandowski. 2000. Mapping of the tobacco mosaic virus movement protein and coat protein subgenomic RNA promoters in vivo. Virology 275:177-192. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

