Acetylated Tat regulates human immunodeficiency virus type 1 splicing through its interaction with the splicing regulator p32
- PMID: 16537587
- PMCID: PMC1440361
- DOI: 10.1128/JVI.80.7.3189-3204.2006
Acetylated Tat regulates human immunodeficiency virus type 1 splicing through its interaction with the splicing regulator p32
Abstract
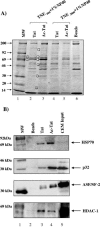
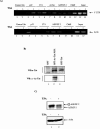
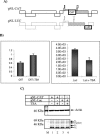
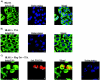
The human immunodeficiency virus type 1 (HIV-1) potent transactivator Tat protein mediates pleiotropic effects on various cell functions. Posttranslational modification of Tat affects its activity during viral transcription. Tat binds to TAR and subsequently becomes acetylated on lysine residues by histone acetyltransferases. Novel protein-protein interaction domains on acetylated Tat are then established, which are necessary for both sustained transcriptional activation of the HIV-1 promoter and viral transcription elongation. In this study, we investigated the identity of proteins that preferentially bound acetylated Tat. Using a proteomic approach, we identified a number of proteins that preferentially bound AcTat, among which p32, a cofactor of splicing factor ASF/SF-2, was identified. We found that p32 was recruited to the HIV-1 genome, suggesting a mechanism by which acetylation of Tat may inhibit HIV-1 splicing needed for the production of full-length transcripts. Using Tat from different clades, harboring a different number of acetylation sites, as well as Tat mutated at lysine residues, we demonstrated that Tat acetylation affected splicing in vivo. Finally, using confocal microscopy, we found that p32 and Tat colocalize in vivo in HIV-1-infected cells.
Figures






Similar articles
-
HIV-1 tat transcriptional activity is regulated by acetylation.EMBO J. 1999 Nov 1;18(21):6106-18. doi: 10.1093/emboj/18.21.6106. EMBO J. 1999. PMID: 10545121 Free PMC article.
-
Tat acetylation: a regulatory switch between early and late phases in HIV transcription elongation.Novartis Found Symp. 2004;259:182-93; discussion 193-6, 223-5. Novartis Found Symp. 2004. PMID: 15171254 Review.
-
Acetylation of HIV-1 Tat by CBP/P300 increases transcription of integrated HIV-1 genome and enhances binding to core histones.Virology. 2000 Nov 25;277(2):278-95. doi: 10.1006/viro.2000.0593. Virology. 2000. PMID: 11080476
-
Effect of SWI/SNF chromatin remodeling complex on HIV-1 Tat activated transcription.Retrovirology. 2006 Aug 7;3:48. doi: 10.1186/1742-4690-3-48. Retrovirology. 2006. PMID: 16893449 Free PMC article.
-
Multiple modes of transcriptional regulation by the HIV-1 Tat transactivator.IUBMB Life. 2001 Mar;51(3):175-81. doi: 10.1080/152165401753544241. IUBMB Life. 2001. PMID: 11547919 Review.
Cited by
-
Lost in transcription: molecular mechanisms that control HIV latency.Viruses. 2013 Mar 21;5(3):902-27. doi: 10.3390/v5030902. Viruses. 2013. PMID: 23518577 Free PMC article. Review.
-
DC-SIGN, C1q, and gC1qR form a trimolecular receptor complex on the surface of monocyte-derived immature dendritic cells.Blood. 2012 Aug 9;120(6):1228-36. doi: 10.1182/blood-2011-07-369728. Epub 2012 Jun 13. Blood. 2012. PMID: 22700724 Free PMC article.
-
The chaperone protein p32 stabilizes HIV-1 Tat and strengthens the p-TEFb/RNAPII/TAR complex promoting HIV transcription elongation.Proc Natl Acad Sci U S A. 2023 Jan 3;120(1):e2217476120. doi: 10.1073/pnas.2217476120. Epub 2022 Dec 30. Proc Natl Acad Sci U S A. 2023. PMID: 36584296 Free PMC article.
-
Role of Host Cell p32 in Herpes Simplex Virus 1 De-Envelopment during Viral Nuclear Egress.J Virol. 2015 Sep;89(17):8982-98. doi: 10.1128/JVI.01220-15. Epub 2015 Jun 17. J Virol. 2015. PMID: 26085152 Free PMC article.
-
Structure of the Trypanosoma brucei p22 protein, a cytochrome oxidase subunit II-specific RNA-editing accessory factor.J Biol Chem. 2010 Jun 11;285(24):18899-908. doi: 10.1074/jbc.M109.066597. Epub 2010 Apr 14. J Biol Chem. 2010. PMID: 20392699 Free PMC article.
References
-
- Adair, R., G. W. Liebisch, Y. Su, and A. M. Colberg-Poley. 2004. Alteration of cellular RNA splicing and polyadenylation machineries during productive human cytomegalovirus infection. J. Gen. Virol. 85:3541-3553. - PubMed
-
- Balasubramanyam, K., R. Varier, M. Altaf, V. Swaminathan, N. Siddappa, U. Ranga, and T. Kundu. 2004. Curcumin, a novel p300/CREB-binding protein-specific inhibitor of acetyltransferase, represses the acetylation of histone/nonhistone proteins and histone acetyltransferase-dependent chromatin transcription. J. Biol. Chem. 279:51163-51171. - PubMed
-
- Benkirane, M., R. F. Chun, H. Xiao, V. V. Ogryzko, B. H. Howard, Y. Nakatani, and K. T. Jeang. 1998. Activation of integrated provirus requires histone acetyltransferase. p300 and P/CAF are coactivators for HIV-1 Tat. J. Biol. Chem. 273:24898-24905. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

