T-bet regulates Th1 responses through essential effects on GATA-3 function rather than on IFNG gene acetylation and transcription
- PMID: 16520391
- PMCID: PMC2118252
- DOI: 10.1084/jem.20052165
T-bet regulates Th1 responses through essential effects on GATA-3 function rather than on IFNG gene acetylation and transcription
Erratum in
- J Exp Med. 2006 Apr 17;203(4):1129
Abstract
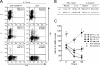
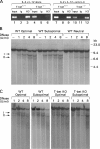
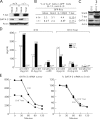
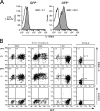
T helper type 1 (Th1) development is facilitated by interrelated changes in key intracellular factors, particularly signal transducer and activator of transcription (STAT)4, T-bet, and GATA-3. Here we show that CD4+ cells from T-bet-/- mice are skewed toward Th2 differentiation by high endogenous GATA-3 levels but exhibit virtually normal Th1 differentiation provided that GATA-3 levels are regulated at an early stage by anti-interleukin (IL)-4 blockade of IL-4 receptor (R) signaling. In addition, under these conditions, Th1 cells from T-bet-/- mice manifest IFNG promotor accessibility as detected by histone acetylation and deoxyribonuclease I hypersensitivity. In related studies, we show that the negative effect of GATA-3 on Th1 differentiation in T-bet-/- cells arises from its ability to suppress STAT4 levels, because if this is prevented by a STAT4-expressing retrovirus, normal Th1 differentiation is observed. Finally, we show that retroviral T-bet expression in developing and established Th2 cells leads to down-regulation of GATA-3 levels. These findings lead to a model of T cell differentiation that holds that naive T cells tend toward Th2 differentiation through induction of GATA-3 and subsequent down-regulation of STAT4/IL-12Rbeta2 chain unless GATA-3 levels or function is regulated by T-bet. Thus, the principal function of T-bet in developing Th1 cells is to negatively regulate GATA-3 rather than to positively regulate the IFNG gene.
Figures



Similar articles
-
[Imbalanced T cell-specific transcription factors T-bet and GATA-3 contributes to type 2 T helper cell polarization in asthmatic patients].Zhonghua Jie He He Hu Xi Za Zhi. 2004 Jun;27(6):398-402. Zhonghua Jie He He Hu Xi Za Zhi. 2004. PMID: 15256090 Chinese.
-
Elevated gene expression of Th1/Th2 associated transcription factors is correlated with disease activity in patients with systemic lupus erythematosus.J Rheumatol. 2007 Jan;34(1):89-96. Epub 2006 Nov 15. J Rheumatol. 2007. PMID: 17117487
-
T-bet is a STAT1-induced regulator of IL-12R expression in naïve CD4+ T cells.Nat Immunol. 2002 Jun;3(6):549-57. doi: 10.1038/ni794. Epub 2002 May 13. Nat Immunol. 2002. PMID: 12006974
-
GATA-3 promotes Th2 responses through three different mechanisms: induction of Th2 cytokine production, selective growth of Th2 cells and inhibition of Th1 cell-specific factors.Cell Res. 2006 Jan;16(1):3-10. doi: 10.1038/sj.cr.7310002. Cell Res. 2006. PMID: 16467870 Review.
-
Chromatin remodeling and T helper subset differentiation.IUBMB Life. 2000 Jun;49(6):473-8. doi: 10.1080/15216540050166990. IUBMB Life. 2000. PMID: 11032239 Review.
Cited by
-
Proviral integration site for Moloney murine leukemia virus (PIM) kinases promote human T helper 1 cell differentiation.J Biol Chem. 2013 Feb 1;288(5):3048-58. doi: 10.1074/jbc.M112.361709. Epub 2012 Dec 3. J Biol Chem. 2013. PMID: 23209281 Free PMC article.
-
Deficiency in Th2 cytokine responses exacerbate orthopoxvirus infection.PLoS One. 2015 Mar 9;10(3):e0118685. doi: 10.1371/journal.pone.0118685. eCollection 2015. PLoS One. 2015. PMID: 25751266 Free PMC article.
-
Essentials of Th17 cell commitment and plasticity.Blood. 2013 Mar 28;121(13):2402-14. doi: 10.1182/blood-2012-09-378653. Epub 2013 Jan 16. Blood. 2013. PMID: 23325835 Free PMC article. Review.
-
Normal and cancer fibroblasts differentially regulate TWIST1, TOX and cytokine gene expression in cutaneous T-cell lymphoma.BMC Cancer. 2021 May 3;21(1):492. doi: 10.1186/s12885-021-08142-7. BMC Cancer. 2021. PMID: 33941102 Free PMC article.
-
Helper T cell diversity and plasticity.Curr Opin Immunol. 2012 Jun;24(3):297-302. doi: 10.1016/j.coi.2012.01.014. Epub 2012 Feb 15. Curr Opin Immunol. 2012. PMID: 22341735 Free PMC article. Review.
References
-
- Hsieh, C.S., S.E. Macatonia, C.S. Tripp, S.F. Wolf, A. O'Garra, and K.M. Murphy. 1993. Development of Th1 CD4+ T cells through IL-12 produced by Listeria-induced macrophages. Science. 260:547–549. - PubMed
-
- Jacobson, N.G., S.J. Szabo, R.M. Weber-Nordt, Z. Zhong, R.D. Schreiber, J.E. Darnell Jr., and K.M. Murphy. 1995. Interleukin 12 signaling in T helper type 1 (Th1) cells involves tyrosine phosphorylation of signal transducer and activator of transcription (STAT)3 and STAT 4. J. Exp. Med. 181:1755–1762. - PMC - PubMed
-
- Kaplan, M.H., Y.L. Sun, T. Hoey, and J.M. Grusby. 1996. Impaired IL-12 responses and enhanced development of Th2 cells in STAT4-deficient mice. Nature. 382:174–177. - PubMed
-
- Magram, J., S.E. Connaughton, R.R. Warrier, D.M. Carvajal, C.Y. Wu, J. Ferrante, C. Stewart, U. Sarmiento, D.A. Faherty, and M.K. Gately. 1996. IL-12-deficient mice are defective in IFN-γ production and type 1 cytokine responses. Immunity. 4:471–481. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

