Schizosaccharomyces pombe Git1 is a C2-domain protein required for glucose activation of adenylate cyclase
- PMID: 16489217
- PMCID: PMC1461440
- DOI: 10.1534/genetics.106.055699
Schizosaccharomyces pombe Git1 is a C2-domain protein required for glucose activation of adenylate cyclase
Abstract
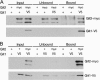
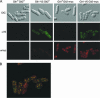
Schizosaccharomyces pombe senses environmental glucose through a cAMP-signaling pathway, activating cAMP-dependent protein kinase A (PKA). This requires nine git (glucose insensitive transcription) genes that encode adenylate cyclase, the PKA catalytic subunit, and seven "upstream" proteins required for glucose-triggered adenylate cyclase activation, including three heterotrimeric G-protein subunits and its associated receptor. We describe here the cloning and characterization of the git1+ gene. Git1 is distantly related to a small group of uncharacterized fungal proteins, including a second S. pombe protein that is not functionally redundant with Git1, as well as to members of the UNC-13/Munc13 protein family. Mutations in git1+ demonstrate functional roles for the two most highly conserved regions of the protein, the C2 domain and the MHD2 Munc homology domain. Cells lacking Git1 are viable, but display phenotypes associated with cAMP-signaling defects, even in strains expressing a mutationally activated G alpha-subunit, which activates adenylate cyclase. These cells possess reduced basal cAMP levels and fail to mount a cAMP response to glucose. In addition, Git1 and adenylate cyclase physically interact and partially colocalize in the cell. Thus, Git1 is a critical component of the S. pombe glucose/cAMP pathway.
Figures







Similar articles
-
Schizosaccharomyces pombe Hsp90/Git10 is required for glucose/cAMP signaling.Genetics. 2008 Apr;178(4):1927-36. doi: 10.1534/genetics.107.086165. Genetics. 2008. PMID: 18430926 Free PMC article.
-
Schizosaccharomyces pombe Git7p, a member of the Saccharomyces cerevisiae Sgtlp family, is required for glucose and cyclic AMP signaling, cell wall integrity, and septation.Eukaryot Cell. 2002 Aug;1(4):558-67. doi: 10.1128/EC.1.4.558-567.2002. Eukaryot Cell. 2002. PMID: 12456004 Free PMC article.
-
Glucose monitoring in fission yeast via the Gpa2 galpha, the git5 Gbeta and the git3 putative glucose receptor.Genetics. 2000 Oct;156(2):513-21. doi: 10.1093/genetics/156.2.513. Genetics. 2000. PMID: 11014802 Free PMC article.
-
Glucose sensing via the protein kinase A pathway in Schizosaccharomyces pombe.Biochem Soc Trans. 2005 Feb;33(Pt 1):257-60. doi: 10.1042/BST0330257. Biochem Soc Trans. 2005. PMID: 15667320 Free PMC article. Review.
-
Regulation of sexual differentiation initiation in Schizosaccharomyces pombe.Biosci Biotechnol Biochem. 2024 Apr 22;88(5):475-492. doi: 10.1093/bbb/zbae019. Biosci Biotechnol Biochem. 2024. PMID: 38449372 Review.
Cited by
-
A fission yeast-based platform for phosphodiesterase inhibitor HTSs and analyses of phosphodiesterase activity.Handb Exp Pharmacol. 2011;(204):135-49. doi: 10.1007/978-3-642-17969-3_5. Handb Exp Pharmacol. 2011. PMID: 21695638 Free PMC article. Review.
-
Schizosaccharomyces pombe Hsp90/Git10 is required for glucose/cAMP signaling.Genetics. 2008 Apr;178(4):1927-36. doi: 10.1534/genetics.107.086165. Genetics. 2008. PMID: 18430926 Free PMC article.
-
Roles of the fission yeast UNC-13/Munc13 protein Ync13 in late stages of cytokinesis.Mol Biol Cell. 2018 Sep 15;29(19):2259-2279. doi: 10.1091/mbc.E18-04-0225. Epub 2018 Jul 25. Mol Biol Cell. 2018. PMID: 30044717 Free PMC article.
-
Multistep regulation of protein kinase A in its localization, phosphorylation and binding with a regulatory subunit in fission yeast.Curr Genet. 2011 Oct;57(5):353-65. doi: 10.1007/s00294-011-0354-2. Epub 2011 Aug 31. Curr Genet. 2011. PMID: 21879336
-
Cellular stress induces cytoplasmic RNA granules in fission yeast.RNA. 2011 Jan;17(1):120-33. doi: 10.1261/rna.2268111. Epub 2010 Nov 22. RNA. 2011. PMID: 21098141 Free PMC article.
References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers and D. J. Lipman, 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. - PubMed
-
- Bähler, J., J. Q. Wu, M. S. Longtine, N. G. Shah, A. McKenzie, 3rd et al., 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14: 943–951. - PubMed
-
- Batlle, M., A. Lu, D. A. Green, Y. Xue and J. P. Hirsch, 2003. Krh1p and Krh2p act downstream of the Gpa2p Gα subunit to negatively regulate haploid invasive growth. J. Cell Sci. 116: 701–710. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

