A chromatin insulator-like element in the herpes simplex virus type 1 latency-associated transcript region binds CCCTC-binding factor and displays enhancer-blocking and silencing activities
- PMID: 16474142
- PMCID: PMC1395413
- DOI: 10.1128/JVI.80.5.2358-2368.2006
A chromatin insulator-like element in the herpes simplex virus type 1 latency-associated transcript region binds CCCTC-binding factor and displays enhancer-blocking and silencing activities
Abstract
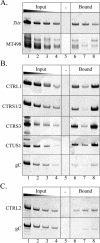
A previous study demonstrated that the latency-associated transcript (LAT) promoter and the LAT enhancer/reactivation critical region (rcr) are enriched in acetyl histone H3 (K9, K14) during herpes simplex virus type 1 (HSV-1) latency, whereas all lytic genes analyzed (ICP0, UL54, ICP4, and DNA polymerase) are not (N. J. Kubat, R. K. Tran, P. McAnany, and D. C. Bloom, J. Virol. 78:1139-1149, 2004). This suggests that the HSV-1 latent genome is organized into histone H3 (K9, K14) hyperacetylated and hypoacetylated regions corresponding to transcriptionally permissive and transcriptionally repressed chromatin domains, respectively. Such an organization implies that chromatin insulators, similar to those of cellular chromosomes, may separate distinct transcriptional domains of the HSV-1 latent genome. In the present study, we sought to identify cis elements that could partition the HSV-1 genome into distinct chromatin domains. Sequence analysis coupled with chromatin immunoprecipitation and luciferase reporter assays revealed that (i) the long and short repeats and the unique-short region of the HSV-1 genome contain clustered CTCF (CCCTC-binding factor) motifs, (ii) CTCF motif clusters similar to those in HSV-1 are conserved in other alphaherpesviruses, (iii) CTCF binds to these motifs on latent HSV-1 genomes in vivo, and (iv) a 1.5-kb region containing the CTCF motif cluster in the LAT region possesses insulator activities, specifically, enhancer blocking and silencing. The finding that CTCF, a cellular protein associated with chromatin insulators, binds to motifs on the latent genome and insulates the LAT enhancer suggests that CTCF may facilitate the formation of distinct chromatin boundaries during herpesvirus latency.
Figures




Similar articles
-
CTCF-dependent chromatin boundary element between the latency-associated transcript and ICP0 promoters in the herpes simplex virus type 1 genome.J Virol. 2007 May;81(10):5192-201. doi: 10.1128/JVI.02447-06. Epub 2007 Jan 31. J Virol. 2007. PMID: 17267480 Free PMC article.
-
CTCF Binding Sites in the Herpes Simplex Virus 1 Genome Display Site-Specific CTCF Occupation, Protein Recruitment, and Insulator Function.J Virol. 2018 Mar 28;92(8):e00156-18. doi: 10.1128/JVI.00156-18. Print 2018 Apr 15. J Virol. 2018. PMID: 29437965 Free PMC article.
-
The CCCTC Binding Factor, CTRL2, Modulates Heterochromatin Deposition and the Establishment of Herpes Simplex Virus 1 Latency In Vivo.J Virol. 2019 Jun 14;93(13):e00415-19. doi: 10.1128/JVI.00415-19. Print 2019 Jul 1. J Virol. 2019. PMID: 30996085 Free PMC article.
-
Epigenetic regulation of latent HSV-1 gene expression.Biochim Biophys Acta. 2010 Mar-Apr;1799(3-4):246-56. doi: 10.1016/j.bbagrm.2009.12.001. Epub 2010 Jan 4. Biochim Biophys Acta. 2010. PMID: 20045093 Free PMC article. Review.
-
Insulators and domains of gene expression.Curr Opin Genet Dev. 2016 Apr;37:17-26. doi: 10.1016/j.gde.2015.11.009. Epub 2016 Jan 20. Curr Opin Genet Dev. 2016. PMID: 26802288 Review.
Cited by
-
CTCF occupation of the herpes simplex virus 1 genome is disrupted at early times postreactivation in a transcription-dependent manner.J Virol. 2012 Dec;86(23):12741-59. doi: 10.1128/JVI.01655-12. Epub 2012 Sep 12. J Virol. 2012. PMID: 22973047 Free PMC article.
-
Effects of thyroid hormone on HSV-1 gene regulation: implications in the control of viral latency and reactivation.Cell Biosci. 2011 Jul 14;1(1):24. doi: 10.1186/2045-3701-1-24. Cell Biosci. 2011. PMID: 21756309 Free PMC article.
-
The critical role of human transcriptional repressor CTCF mRNA up-regulation in the induction of anti-HIV-1 responses in CD4(+) T cells.Immunol Lett. 2008 Apr 15;117(1):35-44. doi: 10.1016/j.imlet.2007.11.017. Epub 2007 Dec 26. Immunol Lett. 2008. PMID: 18207574 Free PMC article.
-
CCCTC-Binding Factor Acts as a Heterochromatin Barrier on Herpes Simplex Viral Latent Chromatin and Contributes to Poised Latent Infection.mBio. 2018 Feb 6;9(1):e02372-17. doi: 10.1128/mBio.02372-17. mBio. 2018. PMID: 29437926 Free PMC article.
-
Genital Herpes: Insights into Sexually Transmitted Infectious Disease.Microb Cell. 2016 Jun 27;3(9):438-450. doi: 10.15698/mic2016.09.528. Microb Cell. 2016. PMID: 28357380 Free PMC article. Review.
References
-
- Bell, A. C., A. G. West, and G. Felsenfeld. 1999. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98:387-396. - PubMed
-
- Burcin, M., R. Arnold, M. Lutz, B. Kaiser, D. Runge, F. Lottspeich, G. N. Filippova, V. V. Lobanenkov, and R. Renkawitz. 1997. Negative protein 1, which is required for function of the chicken lysozyme gene silencer in conjunction with hormone receptors, is identical to the multivalent zinc finger repressor CTCF. Mol. Cell. Biol. 17:1281-1288. - PMC - PubMed
-
- Capelson, M., and V. G. Corces. 2004. Boundary elements and nuclear organization. Biol. Cell 96:617-629. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

