Forced-unfolding and force-quench refolding of RNA hairpins
- PMID: 16473903
- PMCID: PMC1440726
- DOI: 10.1529/biophysj.105.078030
Forced-unfolding and force-quench refolding of RNA hairpins
Abstract
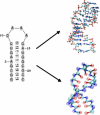
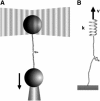
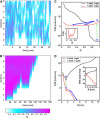
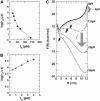
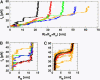
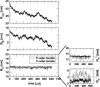
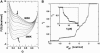
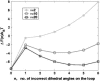
Nanomanipulation of individual RNA molecules, using laser optical tweezers, has made it possible to infer the major features of their energy landscape. Time-dependent mechanical unfolding trajectories, measured at a constant stretching force (f(S)) of simple RNA structures (hairpins and three-helix junctions) sandwiched between RNA/DNA hybrid handles show that they unfold in a reversible all-or-none manner. To provide a molecular interpretation of the experiments we use a general coarse-grained off-lattice Gō-like model, in which each nucleotide is represented using three interaction sites. Using the coarse-grained model we have explored forced-unfolding of RNA hairpin as a function of f(S) and the loading rate (r(f)). The simulations and theoretical analysis have been done both with and without the handles that are explicitly modeled by semiflexible polymer chains. The mechanisms and timescales for denaturation by temperature jump and mechanical unfolding are vastly different. The directed perturbation of the native state by f(S) results in a sequential unfolding of the hairpin starting from their ends, whereas thermal denaturation occurs stochastically. From the dependence of the unfolding rates on r(f) and f(S) we show that the position of the unfolding transition state is not a constant but moves dramatically as either r(f) or f(S) is changed. The transition-state movements are interpreted by adopting the Hammond postulate for forced-unfolding. Forced-unfolding simulations of RNA, with handles attached to the two ends, show that the value of the unfolding force increases (especially at high pulling speeds) as the length of the handles increases. The pathways for refolding of RNA from stretched initial conformation, upon quenching f(S) to the quench force f(Q), are highly heterogeneous. The refolding times, upon force-quench, are at least an order-of-magnitude greater than those obtained by temperature-quench. The long f(Q)-dependent refolding times starting from fully stretched states are analyzed using a model that accounts for the microscopic steps in the rate-limiting step, which involves the trans to gauche transitions of the dihedral angles in the GAAA tetraloop. The simulations with explicit molecular model for the handles show that the dynamics of force-quench refolding is strongly dependent on the interplay of their contour length and persistence length and the RNA persistence length. Using the generality of our results, we also make a number of precise experimentally testable predictions.
Figures



Similar articles
-
Mechanical unfolding of RNA: from hairpins to structures with internal multiloops.Biophys J. 2007 Feb 1;92(3):731-43. doi: 10.1529/biophysj.106.093062. Epub 2006 Oct 6. Biophys J. 2007. PMID: 17028142 Free PMC article.
-
Mechanical unfolding of RNA hairpins.Proc Natl Acad Sci U S A. 2005 May 10;102(19):6789-94. doi: 10.1073/pnas.0408314102. Epub 2005 Mar 4. Proc Natl Acad Sci U S A. 2005. PMID: 15749822 Free PMC article.
-
Force-dependent fragility in RNA hairpins.Phys Rev Lett. 2006 Jun 2;96(21):218301. doi: 10.1103/PhysRevLett.96.218301. Epub 2006 May 31. Phys Rev Lett. 2006. PMID: 16803276
-
Determination of thermodynamics and kinetics of RNA reactions by force.Q Rev Biophys. 2006 Nov;39(4):325-60. doi: 10.1017/S0033583506004446. Epub 2006 Oct 16. Q Rev Biophys. 2006. PMID: 17040613 Free PMC article. Review.
-
Force as a useful variable in reactions: unfolding RNA.Annu Rev Biophys Biomol Struct. 2004;33:363-85. doi: 10.1146/annurev.biophys.33.110502.140418. Annu Rev Biophys Biomol Struct. 2004. PMID: 15139818 Free PMC article. Review.
Cited by
-
Interplay between Viscoelasticity and Force Rate Affects Sequential Unfolding in Polyproteins Pulled at Constant Velocity.Macromolecules. 2020 Apr 28;53(8):3021-3029. doi: 10.1021/acs.macromol.0c00278. Epub 2020 Apr 14. Macromolecules. 2020. PMID: 32905266 Free PMC article.
-
RNA under tension: Folding Landscapes, Kinetic partitioning Mechanism, and Molecular Tensegrity.J Phys Chem Lett. 2012 Nov 19;3(23):3616-3625. doi: 10.1021/jz301537t. J Phys Chem Lett. 2012. PMID: 23336034 Free PMC article.
-
Folding and unfolding single RNA molecules under tension.Curr Opin Chem Biol. 2008 Dec;12(6):640-6. doi: 10.1016/j.cbpa.2008.08.011. Epub 2008 Sep 9. Curr Opin Chem Biol. 2008. PMID: 18786653 Free PMC article. Review.
-
Hairpins under tension: RNA versus DNA.Nucleic Acids Res. 2015 Nov 16;43(20):9928-36. doi: 10.1093/nar/gkv860. Epub 2015 Aug 31. Nucleic Acids Res. 2015. PMID: 26323319 Free PMC article.
-
Multiple barriers in forced rupture of protein complexes.J Chem Phys. 2012 Aug 7;137(5):055103. doi: 10.1063/1.4739747. J Chem Phys. 2012. PMID: 22894385 Free PMC article.
References
-
- Doudna, J., and T. Cech. 2002. The chemical repertoire of natural ribozymes. Nature. 418:222–228. - PubMed
-
- Onoa, B., and I. Tinoco, Jr. 2004. RNA folding and unfolding. Curr. Opin. Struct. Biol. 14:374–379. - PubMed
-
- Bustamante, C., Y. R. Chemla, N. R. Forde, and D. Izhaky. 2004. Mechanical processes in biochemistry. Annu. Rev. Biochem. 73:705–748. - PubMed
-
- Liphardt, J., B. Onoa, S. B. Smith, I. Tinoco, Jr., and C. Bustamante. 2001. Reversible unfolding of single RNA molecules by mechanical force. Science. 292:733–737. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

