Complex interactions of HIV-1 nucleocapsid protein with oligonucleotides
- PMID: 16434700
- PMCID: PMC1351370
- DOI: 10.1093/nar/gkj442
Complex interactions of HIV-1 nucleocapsid protein with oligonucleotides
Erratum in
- Nucleic Acids Res. 2006;34(3):1082
Abstract
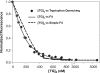
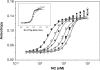
The HIV-1 nucleocapsid (NC) protein is a small, basic protein containing two retroviral zinc fingers. It is a highly active nucleic acid chaperone; because of this activity, it plays a crucial role in virus replication as a cofactor during reverse transcription, and is probably important in other steps of the replication cycle as well. We previously reported that NC binds with high-affinity to the repeating sequence d(TG)n. We have now analyzed the interaction between NC and d(TG)4 in considerable detail, using surface plasmon resonance (SPR), tryptophan fluorescence quenching (TFQ), fluorescence anisotropy (FA), isothermal titration calorimetry (ITC) and electrospray ionization Fourier transform mass spectrometry (ESI-FTMS). Our results show that the interactions between these two molecules are surprisngly complex: while the K(d) for binding of a single d(TG)4 molecule to NC is only approximately 5 nM in 150 mM NaCl, a single NC molecule is capable of interacting with more than one d(TG)4 molecule, and conversely, more than one NC molecule can bind to a single d(TG)4 molecule. The strengths of these additional binding reactions are quantitated. The implications of this multivalency for the functions of NC in virus replication are discussed.
Figures


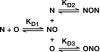
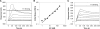


Similar articles
-
Sequence-specific binding of human immunodeficiency virus type 1 nucleocapsid protein to short oligonucleotides.J Virol. 1998 Mar;72(3):1902-9. doi: 10.1128/JVI.72.3.1902-1909.1998. J Virol. 1998. PMID: 9499042 Free PMC article.
-
Structural determinants of HIV-1 nucleocapsid protein for cTAR DNA binding and destabilization, and correlation with inhibition of self-primed DNA synthesis.J Mol Biol. 2005 May 20;348(5):1113-26. doi: 10.1016/j.jmb.2005.02.042. Epub 2005 Apr 1. J Mol Biol. 2005. PMID: 15854648
-
Inhibitory effects of archetypical nucleic acid ligands on the interactions of HIV-1 nucleocapsid protein with elements of Psi-RNA.Nucleic Acids Res. 2006 Mar 6;34(5):1305-16. doi: 10.1093/nar/gkl004. Print 2006. Nucleic Acids Res. 2006. PMID: 16522643 Free PMC article.
-
Methods for the analysis of HIV-1 nucleocapsid protein interactions with oligonucleotides.Methods Mol Biol. 2009;485:209-21. doi: 10.1007/978-1-59745-170-3_15. Methods Mol Biol. 2009. PMID: 19020828
-
Direct mass spectrometric determination of the stoichiometry and binding affinity of the complexes between nucleocapsid protein and RNA stem-loop hairpins of the HIV-1 Psi-recognition element.Biochemistry. 2003 Sep 16;42(36):10736-45. doi: 10.1021/bi0348922. Biochemistry. 2003. PMID: 12962498
Cited by
-
Mechanisms and inhibition of HIV integration.Drug Discov Today Dis Mech. 2006 Jul 1;3(2):253-260. doi: 10.1016/j.ddmec.2006.05.004. Drug Discov Today Dis Mech. 2006. PMID: 20431697 Free PMC article.
-
The DNA binding and 3'-end preferential activity of human tyrosyl-DNA phosphodiesterase.Nucleic Acids Res. 2010 Apr;38(7):2444-52. doi: 10.1093/nar/gkp1206. Epub 2010 Jan 21. Nucleic Acids Res. 2010. PMID: 20097655 Free PMC article.
-
Dissecting the oligonucleotide binding properties of a disordered chaperone protein using surface plasmon resonance.Nucleic Acids Res. 2013 Dec;41(22):10414-25. doi: 10.1093/nar/gkt792. Epub 2013 Sep 11. Nucleic Acids Res. 2013. PMID: 24030713 Free PMC article.
-
Bifunctional cross-linking approaches for mass spectrometry-based investigation of nucleic acids and protein-nucleic acid assemblies.Methods. 2018 Jul 15;144:64-78. doi: 10.1016/j.ymeth.2018.05.001. Epub 2018 May 10. Methods. 2018. PMID: 29753003 Free PMC article.
-
The HIV-1 nucleocapsid protein recruits negatively charged lipids to ensure its optimal binding to lipid membranes.J Virol. 2015 Feb;89(3):1756-67. doi: 10.1128/JVI.02931-14. Epub 2014 Nov 19. J Virol. 2015. PMID: 25410868 Free PMC article.
References
-
- Swanstrom R., Wills J.W. Synthesis, assembly, and processing of viral proteins. In: Coffin J.M., Hughes S.H., Varmus H.E., editors. Retroviruses. Plainview, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 263–334. - PubMed
-
- Berkowitz R., Fisher J., Goff S.P. RNA packaging. Curr. Top Microbiol. Immunol. 1996;214:177–218. - PubMed
-
- Feng Y.X., Campbell S., Harvin D., Ehresmann B., Ehresmann C., Rein A. The human immunodeficiency virus type 1 Gag polyprotein has nucleic acid chaperone activity: possible role in dimerization of genomic RNA and placement of tRNA on the primer binding site. J. Virol. 1999;73:4251–4256. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

