Palmitoylations on murine coronavirus spike proteins are essential for virion assembly and infectivity
- PMID: 16415005
- PMCID: PMC1346925
- DOI: 10.1128/JVI.80.3.1280-1289.2006
Palmitoylations on murine coronavirus spike proteins are essential for virion assembly and infectivity
Abstract
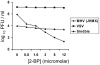
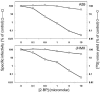
Coronavirus spike (S) proteins are palmitoylated at several cysteine residues clustered near their transmembrane-spanning domains. This is achieved by cellular palmitoyl acyltransferases (PATs), which can modify newly synthesized S proteins before they are assembled into virion envelopes at the intermediate compartment of the exocytic pathway. To address the importance of these fatty acylations to coronavirus infection, we exposed infected cells to 2-bromopalmitate (2-BP), a specific PAT inhibitor. 2-BP profoundly reduced the specific infectivities of murine coronaviruses at very low, nontoxic doses that were inert to alphavirus and rhabdovirus infections. 2-BP effected only two- to fivefold reductions in S palmitoylation, yet this correlated with reduced S complexing with virion membrane (M) proteins and consequent exclusion of S from virions. At defined 2-BP doses, underpalmitoylated S proteins instead trafficked to infected cell surfaces and elicited cell-cell membrane fusions, suggesting that the acyl chain adducts are more critical to virion assembly than to S-induced syncytial developments. These studies involving pharmacologic inhibition of S protein palmitoylation were complemented with molecular genetic analyses in which cysteine acylation substrates were mutated. Notably, some mutations (C1347F and C1348S) did not interfere with S incorporation into virions, indicating that only a subset of the cysteine-rich region provides the essential S-assembly functions. However, the C1347F/C1348S mutant viruses exhibited relatively low specific infectivities, similar to virions secreted from 2-BP-treated cultures. Our collective results indicate that the palmitate adducts on coronavirus S proteins are necessary in assembly and also in positioning the assembled envelope proteins for maximal infectivity.
Figures




Similar articles
-
Genetic analysis of determinants for spike glycoprotein assembly into murine coronavirus virions: distinct roles for charge-rich and cysteine-rich regions of the endodomain.J Virol. 2004 Sep;78(18):9904-17. doi: 10.1128/JVI.78.18.9904-9917.2004. J Virol. 2004. PMID: 15331724 Free PMC article.
-
The viral spike protein is not involved in the polarized sorting of coronaviruses in epithelial cells.J Virol. 1998 Jan;72(1):497-503. doi: 10.1128/JVI.72.1.497-503.1998. J Virol. 1998. PMID: 9420251 Free PMC article.
-
Role of spike protein endodomains in regulating coronavirus entry.J Biol Chem. 2009 Nov 20;284(47):32725-34. doi: 10.1074/jbc.M109.043547. Epub 2009 Sep 30. J Biol Chem. 2009. PMID: 19801669 Free PMC article.
-
Site directed mutagenesis of the murine coronavirus spike protein. Effects on fusion.Adv Exp Med Biol. 1995;380:283-6. doi: 10.1007/978-1-4615-1899-0_45. Adv Exp Med Biol. 1995. PMID: 8830493 Review.
-
[Structure and biological functions of the spike protein of mouse hepatitis virus].Uirusu. 1996 Dec;46(2):109-17. doi: 10.2222/jsv.46.109. Uirusu. 1996. PMID: 9123883 Review. Japanese. No abstract available.
Cited by
-
Topology-Driven Discovery of Transmembrane Protein S-Palmitoylation.bioRxiv [Preprint]. 2024 Sep 8:2024.09.08.611865. doi: 10.1101/2024.09.08.611865. bioRxiv. 2024. PMID: 39282397 Free PMC article. Preprint.
-
Identification of ZDHHC17 as a Potential Drug Target for Swine Acute Diarrhea Syndrome Coronavirus Infection.mBio. 2021 Oct 26;12(5):e0234221. doi: 10.1128/mBio.02342-21. Epub 2021 Oct 26. mBio. 2021. PMID: 34700373 Free PMC article.
-
Review: A systematic review of virus-like particles of coronavirus: Assembly, generation, chimerism and their application in basic research and in the clinic.Int J Biol Macromol. 2022 Mar 1;200:487-497. doi: 10.1016/j.ijbiomac.2022.01.108. Epub 2022 Jan 20. Int J Biol Macromol. 2022. PMID: 35065135 Free PMC article.
-
Biochemical composition, transmission and diagnosis of SARS-CoV-2.Biosci Rep. 2021 Aug 27;41(8):BSR20211238. doi: 10.1042/BSR20211238. Biosci Rep. 2021. PMID: 34291285 Free PMC article. Review.
-
Beyond Shielding: The Roles of Glycans in the SARS-CoV-2 Spike Protein.ACS Cent Sci. 2020 Oct 28;6(10):1722-1734. doi: 10.1021/acscentsci.0c01056. Epub 2020 Sep 23. ACS Cent Sci. 2020. PMID: 33140034 Free PMC article.
References
-
- Arni, S., S. A. Keilbaugh, A. G. Ostermeyer, and D. A. Brown. 1998. Association of GAP-43 with detergent-resistant membranes requires two palmitoylated cysteine residues. J. Biol. Chem. 273:28478-28485. - PubMed
-
- Brown, D. 2002. Structure and function of membrane rafts. Int. J. Med. Microbiol. 291:433-437. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

