Translational resistance of late alphavirus mRNA to eIF2alpha phosphorylation: a strategy to overcome the antiviral effect of protein kinase PKR
- PMID: 16391235
- PMCID: PMC1356103
- DOI: 10.1101/gad.357006
Translational resistance of late alphavirus mRNA to eIF2alpha phosphorylation: a strategy to overcome the antiviral effect of protein kinase PKR
Abstract
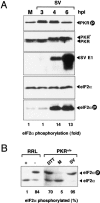
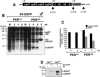
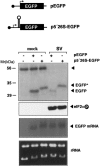
The double-stranded RNA-dependent protein kinase (PKR) is one of the four mammalian kinases that phosphorylates the translation initiation factor 2alpha in response to virus infection. This kinase is induced by interferon and activated by double-stranded RNA (dsRNA). Phosphorylation of eukaryotic initiation factor 2alpha (eIF2alpha) blocks translation initiation of both cellular and viral mRNA, inhibiting virus replication. To counteract this effect, most viruses express inhibitors that prevent PKR activation in infected cells. Here we report that PKR is highly activated following infection with alphaviruses Sindbis (SV) and Semliki Forest virus (SFV), leading to the almost complete phosphorylation of eIF2alpha. Notably, subgenomic SV 26S mRNA is translated efficiently in the presence of phosphorylated eIF2alpha. This modification of eIF2 does not restrict viral replication; SV 26S mRNA initiates translation with canonical methionine in the presence of high levels of phosphorylated eIF2alpha. Genetic and biochemical data showed a highly stable RNA hairpin loop located downstream of the AUG initiator codon that is necessary to provide translational resistance to eIF2alpha phosphorylation. This structure can stall the ribosomes on the correct site to initiate translation of SV 26S mRNA, thus bypassing the requirement for a functional eIF2. Our findings show the existence of an alternative way to locate the ribosomes on the initiation codon of mRNA that is exploited by a family of viruses to counteract the antiviral effect of PKR.
Figures





Similar articles
-
The double-stranded RNA-activated protein kinase PKR is dispensable for regulation of translation initiation in response to either calcium mobilization from the endoplasmic reticulum or essential amino acid starvation.Biochem Biophys Res Commun. 2001 Jan 12;280(1):293-300. doi: 10.1006/bbrc.2000.4103. Biochem Biophys Res Commun. 2001. PMID: 11162513
-
Viral host-range factor C7 or K1 is essential for modified vaccinia virus Ankara late gene expression in human and murine cells, irrespective of their capacity to inhibit protein kinase R-mediated phosphorylation of eukaryotic translation initiation factor 2alpha.J Gen Virol. 2010 Feb;91(Pt 2):470-82. doi: 10.1099/vir.0.015347-0. Epub 2009 Oct 21. J Gen Virol. 2010. PMID: 19846675
-
The Atlantic salmon Z-DNA binding protein kinase phosphorylates translation initiation factor 2 alpha and constitutes a unique orthologue to the mammalian dsRNA-activated protein kinase R.FEBS J. 2008 Jan;275(1):184-97. doi: 10.1111/j.1742-4658.2007.06188.x. Epub 2007 Dec 11. FEBS J. 2008. PMID: 18076653
-
PKR and eIF2alpha: integration of kinase dimerization, activation, and substrate docking.Cell. 2005 Sep 23;122(6):823-5. doi: 10.1016/j.cell.2005.09.007. Cell. 2005. PMID: 16179248 Review.
-
The dsRNA protein kinase PKR: virus and cell control.Biochimie. 2007 Jun-Jul;89(6-7):799-811. doi: 10.1016/j.biochi.2007.03.001. Epub 2007 Mar 12. Biochimie. 2007. PMID: 17451862 Review.
Cited by
-
dsRNA-dependent protein kinase PKR and its role in stress, signaling and HCV infection.Viruses. 2012 Oct 29;4(11):2598-635. doi: 10.3390/v4112598. Viruses. 2012. PMID: 23202496 Free PMC article. Review.
-
ATM kinase is activated by sindbis viral vector infection.Virus Res. 2012 Jun;166(1-2):97-102. doi: 10.1016/j.virusres.2012.03.008. Epub 2012 Mar 29. Virus Res. 2012. PMID: 22475743 Free PMC article.
-
Tinkering with translation: protein synthesis in virus-infected cells.Cold Spring Harb Perspect Biol. 2013 Jan 1;5(1):a012351. doi: 10.1101/cshperspect.a012351. Cold Spring Harb Perspect Biol. 2013. PMID: 23209131 Free PMC article. Review.
-
Roadblocks and fast tracks: How RNA binding proteins affect the viral RNA journey in the cell.Semin Cell Dev Biol. 2021 Mar;111:86-100. doi: 10.1016/j.semcdb.2020.08.006. Epub 2020 Aug 23. Semin Cell Dev Biol. 2021. PMID: 32847707 Free PMC article. Review.
-
PERK Is Critical for Alphavirus Nonstructural Protein Translation.Viruses. 2021 May 12;13(5):892. doi: 10.3390/v13050892. Viruses. 2021. PMID: 34065980 Free PMC article.
References
-
- Adams S.L., Safer, B., Anderson, W.F., and Merrick, W.C. 1975. Eukaryotic initiation complex formation. Evidence for two distinct pathways. J. Biol. Chem. 250: 9083–9089. - PubMed
-
- Anthony D.D. and Merrick, W.C. 1992. Analysis of 40 S and 80 S complexes with mRNA as measured by sucrose density gradients and primer extension inhibition. J. Biol. Chem. 267: 1554–1562. - PubMed
-
- Balachandran S. and Barber, G.N. 2004. Defective translational control facilitates vesicular stomatitis virus oncolysis. Cancer Cell 5: 51–65. - PubMed
-
- Balachandran S., Roberts, P.C., Brown, L.E., Truong, H., Pattnaik, A.K., Archer, D.R., and Barber, G.N. 2000. Essential role for the dsRNA-dependent protein kinase PKR in innate immunity to viral infection. Immunity 13: 129–141. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
