TRANSPATH: an information resource for storing and visualizing signaling pathways and their pathological aberrations
- PMID: 16381929
- PMCID: PMC1347469
- DOI: 10.1093/nar/gkj107
TRANSPATH: an information resource for storing and visualizing signaling pathways and their pathological aberrations
Abstract
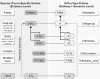
TRANSPATH is a database about signal transduction events. It provides information about signaling molecules, their reactions and the pathways these reactions constitute. The representation of signaling molecules is organized in a number of orthogonal hierarchies reflecting the classification of the molecules, their species-specific or generic features, and their post-translational modifications. Reactions are similarly hierarchically organized in a three-layer architecture, differentiating between reactions that are evidenced by individual publications, generalizations of these reactions to construct species-independent 'reference pathways' and the 'semantic projections' of these pathways. A number of search and browse options allow easy access to the database contents, which can be visualized with the tool PathwayBuildertrade mark. The module PathoSign adds data about pathologically relevant mutations in signaling components, including their genotypes and phenotypes. TRANSPATH and PathoSign can be used as encyclopaedia, in the educational process, for vizualization and modeling of signal transduction networks and for the analysis of gene expression data. TRANSPATH Public 6.0 is freely accessible for users from non-profit organizations under http://www.gene-regulation.com/pub/databases.html.
Figures
Similar articles
-
Integrating pathway data for systems pathology.In Silico Biol. 2007;7(2 Suppl):S17-25. In Silico Biol. 2007. PMID: 17822386
-
TRANSPATH: an integrated database on signal transduction and a tool for array analysis.Nucleic Acids Res. 2003 Jan 1;31(1):97-100. doi: 10.1093/nar/gkg089. Nucleic Acids Res. 2003. PMID: 12519957 Free PMC article.
-
DRUMS: a human disease related unique gene mutation search engine.Hum Mutat. 2011 Oct;32(10):E2259-65. doi: 10.1002/humu.21556. Hum Mutat. 2011. PMID: 21913285
-
FINDbase: a relational database recording frequencies of genetic defects leading to inherited disorders worldwide.Nucleic Acids Res. 2007 Jan;35(Database issue):D690-5. doi: 10.1093/nar/gkl934. Epub 2006 Nov 28. Nucleic Acids Res. 2007. PMID: 17135191 Free PMC article.
-
Mechanisms and causality in molecular diseases.Hist Philos Life Sci. 2017 Oct 16;39(4):35. doi: 10.1007/s40656-017-0162-1. Hist Philos Life Sci. 2017. PMID: 29038918 Free PMC article. Review.
Cited by
-
Systems biology, bioinformatics, and biomarkers in neuropsychiatry.Front Neurosci. 2012 Dec 24;6:187. doi: 10.3389/fnins.2012.00187. eCollection 2012. Front Neurosci. 2012. PMID: 23269912 Free PMC article.
-
Pathway correlation profile of gene-gene co-expression for identifying pathway perturbation.PLoS One. 2012;7(12):e52127. doi: 10.1371/journal.pone.0052127. Epub 2012 Dec 20. PLoS One. 2012. PMID: 23284898 Free PMC article.
-
Mycoplasma hyorhinis reduces sensitivity of human lung carcinoma cells to Nutlin-3 and promotes their malignant phenotype.J Cancer Res Clin Oncol. 2018 Jul;144(7):1289-1300. doi: 10.1007/s00432-018-2658-9. Epub 2018 May 8. J Cancer Res Clin Oncol. 2018. PMID: 29737431
-
Using systems medicine to identify a therapeutic agent with potential for repurposing in inflammatory bowel disease.Dis Model Mech. 2020 Nov 27;13(11):dmm044040. doi: 10.1242/dmm.044040. Dis Model Mech. 2020. PMID: 32958515 Free PMC article.
-
POLAR MAPPER: a computational tool for integrated visualization of protein interaction networks and mRNA expression data.J R Soc Interface. 2009 Oct 6;6(39):881-96. doi: 10.1098/rsif.2008.0407. Epub 2008 Nov 28. J R Soc Interface. 2009. PMID: 19091689 Free PMC article.
References
-
- Takai-Igarashi T., Nadaoka Y., Kaminuma T. A database for cell signaling networks. J. Comput. Biol. 1998;5:747–754. - PubMed
-
- Takai-Igarashi T., Kaminuma T. A pathway finding system for the cell signaling networks database. In Silico Biol. 1999;1:129–146. - PubMed
-
- Schacherer F., Choi C., Götze U., Krull M., Pistor S., Wingender E. The TRANSPATH signal transduction database: a knowledge base on signal transduction networks. Bioinformatics. 2001;17:1053–1057. - PubMed
-
- Demir E., Babur O., Dogrusoz U., Gursoy A., Nisanci G., Cetin-Atalay R., Ozturk M. PATIKA: an integrated visual environment for collaborative construction and analysis of cellular pathways. Bioinformatics. 2002;18:996–1003. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

